8I98
 
 | | Crystal structure of TePixD Y8F | | Descriptor: | FLAVIN MONONUCLEOTIDE, Tll0078 protein | | Authors: | Hu, R, Lin, L, Lu, Q. | | Deposit date: | 2023-02-06 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structure of the BLUF Protein TePixD Y8F Mutant
Progress in Biochemistry and Biophysics, 2023
|
|
8IMS
 
 | | Crystal structure of TRAF7 coiled-coil domain | | Descriptor: | E3 ubiquitin-protein ligase TRAF7 | | Authors: | Hu, R, Lin, L, Lu, Q. | | Deposit date: | 2023-03-07 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structure of TRAF7 coiled-coil trimer provides insight into its function in zebrafish embryonic development.
J Mol Cell Biol, 2024
|
|
8J5Y
 
 | | Structural and mechanistic insight into ribosomal ITS2 RNA processing by nuclease-kinase machinery | | Descriptor: | LAS1 isoform 1, Polynucleotide 5'-hydroxyl-kinase GRC3 | | Authors: | Chen, J, Chen, H, Li, S, Lin, X, Hu, R, Zhang, K, Liu, L. | | Deposit date: | 2023-04-24 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural and mechanistic insights into ribosomal ITS2 RNA processing by nuclease-kinase machinery.
Elife, 12, 2024
|
|
8J60
 
 | | Structural and mechanistic insight into ribosomal ITS2 RNA processing by nuclease-kinase machinery | | Descriptor: | LAS1 protein, Polynucleotide 5'-hydroxyl-kinase GRC3 | | Authors: | Chen, J, Chen, H, Li, S, Lin, X, Hu, R, Zhang, K, Liu, L. | | Deposit date: | 2023-04-24 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural and mechanistic insights into ribosomal ITS2 RNA processing by nuclease-kinase machinery.
Elife, 12, 2024
|
|
6IYN
 
 | | Solution structure of camelid nanobody Nb26 against aflatoxin B1 | | Descriptor: | Nb26 | | Authors: | Nie, Y, He, T, Zhu, J, Li, S.L, Hu, R, Yang, Y.H. | | Deposit date: | 2018-12-17 | | Release date: | 2019-01-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Chemical shift assignments of a camelid nanobody against aflatoxin B1.
Biomol NMR Assign, 13, 2019
|
|
5IT3
 
 | | Swirm domain of human Lsd1 | | Descriptor: | Lysine-specific histone demethylase 1A, MAGNESIUM ION | | Authors: | Jeffrey, P.D, Yuan, P. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Tlx-interacting peptide of Lsd1 inhibits the proliferation of brain tumor stem cells
To Be Published
|
|
6D1W
 
 | | human PKD2 F604P mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Polycystin-2 | | Authors: | Zheng, W, Yang, X, Bulkley, D, Chen, X.Z, Cao, E. | | Deposit date: | 2018-04-12 | | Release date: | 2018-06-27 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Hydrophobic pore gates regulate ion permeation in polycystic kidney disease 2 and 2L1 channels.
Nat Commun, 9, 2018
|
|
6IR2
 
 | | Crystal structure of red fluorescent protein mCherry complexed with the nanobody LaM2 at 1.4 Angstron resolution | | Descriptor: | MCherry fluorescent protein, mCherry's nanobody LaM2 | | Authors: | Ding, Y, Wang, Z.Y, Hu, R.T, Chen, X. | | Deposit date: | 2018-11-09 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.393 Å) | | Cite: | Structural insights into the binding of nanobodies LaM2 and LaM4 to the red fluorescent protein mCherry.
Protein Sci., 30, 2021
|
|
6IR1
 
 | | Crystal structure of red fluorescent protein mCherry complexed with the nanobody LaM4 at 1.9 Angstron resolution | | Descriptor: | MCherry fluorescent protein, mCherry's nanobody LaM4 | | Authors: | Ding, Y, Wang, Z.Y, Hu, R.T, Chen, X. | | Deposit date: | 2018-11-09 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Structural insights into the binding of nanobodies LaM2 and LaM4 to the red fluorescent protein mCherry.
Protein Sci., 30, 2021
|
|
8TJC
 
 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with compound 8a | | Descriptor: | (6M)-1-[(2R)-3,3-dimethylbutan-2-yl]-6-[(5S)-5-methyl-4-oxo-5-phenyl-4,5-dihydro-1H-imidazol-2-yl]-1,3-dihydro-2H-benzimidazol-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sudom, A, Min, X. | | Deposit date: | 2023-07-20 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Imidazolone as an Amide Bioisostere in the Development of beta-1,3- N -Acetylglucosaminyltransferase 2 (B3GNT2) Inhibitors.
J.Med.Chem., 66, 2023
|
|
8SZ3
 
 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with compound 7j | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-[(1S)-1-(5-bromopyridin-2-yl)ethyl]-3-[(2R)-3,3-dimethylbutan-2-yl]-2-oxo-2,3-dihydro-1H-benzimidazole-5-carboxamide, ... | | Authors: | Sudom, A, Min, X. | | Deposit date: | 2023-05-26 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Imidazolone as an Amide Bioisostere in the Development of beta-1,3- N -Acetylglucosaminyltransferase 2 (B3GNT2) Inhibitors.
J.Med.Chem., 66, 2023
|
|
8TIC
 
 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with compound 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-methylpropyl)-2-oxo-2,3-dihydro-1H-benzimidazole-5-carboxylic acid, CHLORIDE ION, ... | | Authors: | Sudom, A, Min, X. | | Deposit date: | 2023-07-19 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Imidazolone as an Amide Bioisostere in the Development of beta-1,3- N -Acetylglucosaminyltransferase 2 (B3GNT2) Inhibitors.
J.Med.Chem., 66, 2023
|
|
8XZF
 
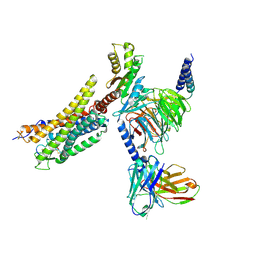 | | Cryo-EM structure of the WN561-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZG
 
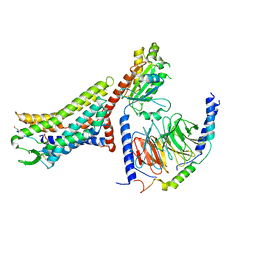 | | Cryo-EM structure of the [Pyr1]-apelin-13-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Apelin-13, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZH
 
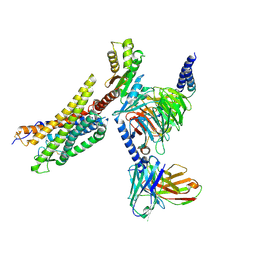 | | Cryo-EM structure of the MM07-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZI
 
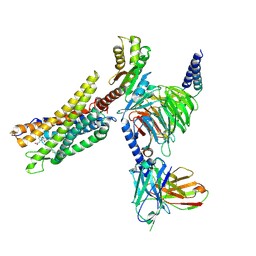 | | Cryo-EM structure of the CMF-019-bound human APLNR-Gi complex | | Descriptor: | (3~{S})-5-methyl-3-[[1-pentan-3-yl-2-(thiophen-2-ylmethyl)benzimidazol-5-yl]carbonylamino]hexanoic acid, Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZJ
 
 | | Cryo-EM structure of the WN353-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
3U78
 
 | | E67-2 selectively inhibits KIAA1718, a human histone H3 lysine 9 Jumonji demethylase | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, 7-[(5-aminopentyl)oxy]-N~4~-(1-benzylpiperidin-4-yl)-N~2~-[3-(dimethylamino)propyl]-6-methoxyquinazoline-2,4-diamine, ... | | Authors: | Upadhyay, A.K, Cheng, X. | | Deposit date: | 2011-10-13 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.689 Å) | | Cite: | An Analog of BIX-01294 Selectively Inhibits a Family of Histone H3 Lysine 9 Jumonji Demethylases.
J.Mol.Biol., 416, 2012
|
|
7F4Q
 
 | | Crystal structure of SAH-bound MTA1-p2 complex | | Descriptor: | MT-a70 family protein, S-ADENOSYL-L-HOMOCYSTEINE, Transmembrane protein, ... | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4O
 
 | | Crystal structure of MTA1-p2 complex | | Descriptor: | MT-a70 family protein, Transmembrane protein, putative | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4T
 
 | |
7F4L
 
 | | Crystal structure of MTA1-p1-p2 complex | | Descriptor: | MT-a70 family protein, Transmembrane protein, putative, ... | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4N
 
 | | Crystal structure of SAH-bound MTA1-p1-p2 complex | | Descriptor: | MT-a70 family protein, S-ADENOSYL-L-HOMOCYSTEINE, Transmembrane protein, ... | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4R
 
 | | Crystal structure of MTA1 | | Descriptor: | MT-a70 family protein | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
7F4S
 
 | | Crystal structure of TthMTA1-PteMTA9 complex | | Descriptor: | MT-a70 family protein, MTA9 | | Authors: | Chen, J, Liu, L. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural basis for MTA1c-mediated DNA N6-adenine methylation
Nat Commun, 13, 2022
|
|
