2FHG
 
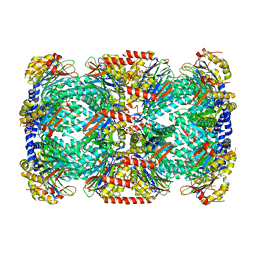 | | Crystal Structure of Mycobacterial Tuberculosis Proteasome | | Descriptor: | 20S proteasome, alpha and beta subunits, proteasome, ... | | Authors: | Hu, G, Lin, G, Wang, M, Dick, L, Xu, R.M, Nathan, C, Li, H. | | Deposit date: | 2005-12-23 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structure of the Mycobacterium tuberculosis proteasome and mechanism of inhibition by a peptidyl boronate.
Mol.Microbiol., 59, 2006
|
|
1JTF
 
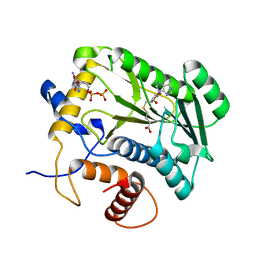 | | Crystal Structure Analysis of VP39-F180W mutant and m7GpppG complex | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Oguro, A, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 2001-08-20 | | Release date: | 2002-07-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The "cap-binding slot" of an mRNA cap-binding protein: quantitative effects of aromatic side chain choice in the double-stacking sandwich with cap.
Biochemistry, 41, 2002
|
|
1JSZ
 
 | | Crystal Structure Analysis of N7,9-dimethylguanine-VP39 complex | | Descriptor: | 7,9-DIMETHYLGUANINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Oguro, A, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 2001-08-19 | | Release date: | 2002-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The "cap-binding slot" of an mRNA cap-binding protein: quantitative effects of aromatic side chain choice in the double-stacking sandwich with cap.
Biochemistry, 41, 2002
|
|
1JTE
 
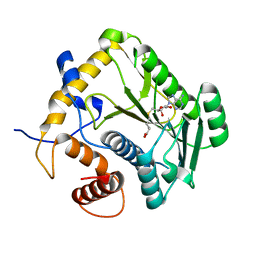 | | Crystal Structure Analysis of VP39 F180W mutant | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Oguro, A, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 2001-08-20 | | Release date: | 2002-07-10 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The "cap-binding slot" of an mRNA cap-binding protein: quantitative effects of aromatic side chain choice in the double-stacking sandwich with cap.
Biochemistry, 41, 2002
|
|
1B42
 
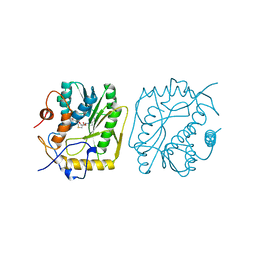 | | VACCINIA METHYLTRANSFERASE VP39 COMPLEXED WITH M1ADE AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 6-AMINO-1-METHYLPURINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Hodel, A.E, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 1999-01-05 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1BKY
 
 | | VACCINIA METHYLTRANSFERASE VP39 COMPLEXED WITH M1CYT AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 1-METHYLCYTOSINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Hodel, A.E, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 1998-07-13 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3JCB
 
 | | Structure of Simian Immunodeficiency Virus Envelope Spikes bound with CD4 and Monoclonal Antibody 36D5 | | Descriptor: | Antibody 36D5 heavy chain, Antibody 36D5 light chain, Envelope glycoprotein gp120, ... | | Authors: | Hu, G, Liu, J, Roux, K, Taylor, K.A. | | Deposit date: | 2015-11-25 | | Release date: | 2017-05-10 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structure of Simian Immunodeficiency Virus Envelope Spikes Bound with CD4 and Monoclonal Antibody 36D5.
J. Virol., 91, 2017
|
|
1EAM
 
 | | VACCINIA METHYLTRANSFERASE VP39 MUTANT (EC: 2.7.7.19) | | Descriptor: | PROTEIN (VP39), S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Hu, G, Hodel, A.E, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 1999-01-26 | | Release date: | 1999-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1EQA
 
 | | VACCINIA METHYLTRANSFERASE VP39 MUTANT E233Q COMPLEXED WITH M7G AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 7-METHYLGUANOSINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Hodel, A.E, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 1999-01-05 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3JCC
 
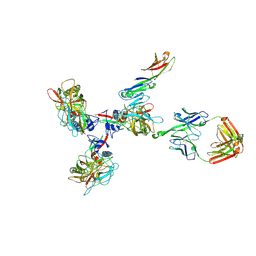 | | Structure of Simian Immunodeficiency Virus Envelope Spikes bound with CD4 and Monoclonal Antibody 36D5 | | Descriptor: | Antibody 36D5 heavy chain, Antibody 36D5 light chain, Envelope glycoprotein gp120, ... | | Authors: | Hu, G, Liu, J, Roux, K, Taylor, K.A. | | Deposit date: | 2015-11-25 | | Release date: | 2017-05-10 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structure of Simian Immunodeficiency Virus Envelope Spikes Bound with CD4 and Monoclonal Antibody 36D5.
J. Virol., 91, 2017
|
|
4DCG
 
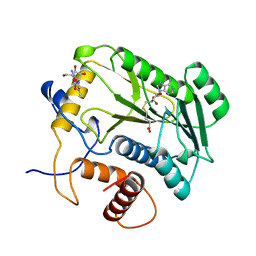 | | VACCINIA METHYLTRANSFERASE VP39 MUTANT D182A COMPLEXED WITH M7G AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 7-METHYLGUANOSINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Hodel, A.E, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 1998-07-13 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3MAG
 
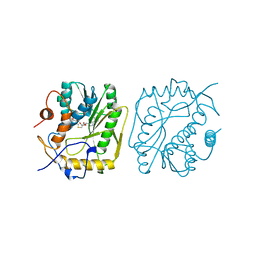 | | VACCINIA METHYLTRANSFERASE VP39 COMPLEXED WITH M3ADE AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 6-AMINO-3-METHYLPURINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Hodel, A.E, Gershon, P.D, Quiocho, F.A. | | Deposit date: | 1998-07-12 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3MCT
 
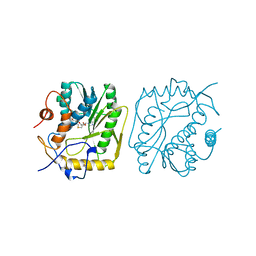 | | VACCINIA METHYLTRANSFERASE VP39 COMPLEXED WITH M3CYT AND S-ADENOSYLHOMOCYSTEINE | | Descriptor: | 3-METHYLCYTOSINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | Authors: | Hu, G, Gershon, P.D, Hodel, A.E, Quiocho, F.A. | | Deposit date: | 1999-01-05 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2H0R
 
 | | Structure of the Yeast Nicotinamidase Pnc1p | | Descriptor: | Nicotinamidase, ZINC ION | | Authors: | Taylor, A.B, Hu, G, Hart, P.J, McAlister-Henn, L. | | Deposit date: | 2006-05-15 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the yeast nicotinamidase Pnc1p.
Arch.Biochem.Biophys., 461, 2007
|
|
1PVZ
 
 | | Solution Structure of BmP07, A Novel Potassium Channel Blocker from Scorpion Buthus martensi Karsch, 15 structures | | Descriptor: | K+ toxin-like peptide | | Authors: | Wu, H, Zhang, N, Wang, Y, Zhang, Q, Ou, L, Li, M, Hu, G. | | Deposit date: | 2003-06-29 | | Release date: | 2004-05-18 | | Last modified: | 2018-06-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKK2, a new potassium channel blocker from the venom of chinese scorpion Buthus martensi Karsch
PROTEINS, 55, 2004
|
|
1WM7
 
 | | Solution Structure of BmP01 from the Venom of Scorpion Buthus martensii Karsch, 9 structures | | Descriptor: | Neurotoxin BmP01 | | Authors: | Wu, G, Li, Y, Wei, D, He, F, Jiang, S, Hu, G, Wu, H, Chen, X. | | Deposit date: | 2004-07-05 | | Release date: | 2004-07-27 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of BmP01 from the Venom of Scorpion Buthus martensii Karsch
Biochem.Biophys.Res.Commun., 276, 2000
|
|
1S8K
 
 | | Solution Structure of BmKK4, A Novel Potassium Channel Blocker from Scorpion Buthus martensii Karsch, 25 structures | | Descriptor: | Toxin BmKK4 | | Authors: | Zhang, N, Chen, X, Li, M, Cao, C, Wang, Y, Hu, G, Wu, H. | | Deposit date: | 2004-02-02 | | Release date: | 2005-02-08 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKK4, the first member of subfamily alpha-KTx 17 of scorpion toxins
Biochemistry, 43, 2004
|
|
3BLX
 
 | | Yeast Isocitrate Dehydrogenase (Apo Form) | | Descriptor: | Isocitrate dehydrogenase [NAD] subunit 1, Isocitrate dehydrogenase [NAD] subunit 2 | | Authors: | Taylor, A.B, Hu, G, Hart, P.J, McAlister-Henn, L. | | Deposit date: | 2007-12-11 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allosteric Motions in Structures of Yeast NAD+-specific Isocitrate Dehydrogenase.
J.Biol.Chem., 283, 2008
|
|
3BLW
 
 | | Yeast Isocitrate Dehydrogenase with Citrate and AMP Bound in the Regulatory Subunits | | Descriptor: | ADENOSINE MONOPHOSPHATE, CITRATE ANION, Isocitrate dehydrogenase [NAD] subunit 1, ... | | Authors: | Taylor, A.B, Hu, G, Hart, P.J, McAlister-Henn, L. | | Deposit date: | 2007-12-11 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Allosteric Motions in Structures of Yeast NAD+-specific Isocitrate Dehydrogenase.
J.Biol.Chem., 283, 2008
|
|
3BLV
 
 | | Yeast Isocitrate Dehydrogenase with Citrate Bound in the Regulatory Subunits | | Descriptor: | CITRATE ANION, Isocitrate dehydrogenase [NAD] subunit 1, Isocitrate dehydrogenase [NAD] subunit 2 | | Authors: | Taylor, A.B, Hu, G, Hart, P.J, McAlister-Henn, L. | | Deposit date: | 2007-12-11 | | Release date: | 2008-02-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Allosteric Motions in Structures of Yeast NAD+-specific Isocitrate Dehydrogenase.
J.Biol.Chem., 283, 2008
|
|
5ZO1
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain (Ig1-Ig3), 2.2A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 4, GLYCEROL | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZO2
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain in complex with mouse nectin-like molecule 1 (mNecl-1) Ig1 domain, 3.3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 3, Cell adhesion molecule 4 | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
2FHH
 
 | |
2VN1
 
 | | Crystal structure of the FK506-binding domain of Plasmodium falciparum FKBP35 in complex with FK506 | | Descriptor: | 70 KDA PEPTIDYLPROLYL ISOMERASE, 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN | | Authors: | Kotaka, M, Alag, R, Ye, H, Preiser, P.R, Yoon, H.S, Lescar, J. | | Deposit date: | 2008-01-30 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the Fk506 Binding Domain of Plasmodium Falciparum Fkbp35 in Complex with Fk506.
Biochemistry, 47, 2008
|
|
1H0D
 
 | | Crystal structure of Human Angiogenin in complex with Fab fragment of its monoclonal antibody mAb 26-2F | | Descriptor: | ANGIOGENIN, ANTIBODY FAB FRAGMENT, HEAVY CHAIN, ... | | Authors: | Chavali, G.B, Papageorgiou, A.C, Acharya, K.R. | | Deposit date: | 2002-06-19 | | Release date: | 2003-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human Angiogenin in Complex with an Antitumor Neutralizing Antibody
Structure, 11, 2003
|
|
