1M8O
 
 | | Platelet integrin alfaIIb-beta3 cytoplasmic domain | | Descriptor: | platelet integrin alfaIIb subunit: cytoplasmic domain, platelet integrin beta3 subunit: cytoplasmic domain | | Authors: | Vinogradova, O, Velyvis, A, Velyviene, A, Hu, B, Haas, T, Plow, E.F, Qin, J. | | Deposit date: | 2002-07-25 | | Release date: | 2002-09-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | A Structural mechanism of integrin alfaiib beta3
"Inside-out" Activation as regulated by its cytoplasmic face.
Cell(Cambridge,Mass.), 110, 2002
|
|
7SPJ
 
 | | Models for C17 reconstruction of Outer Membrane Core Complex (OMCC) of Type IV Secretion System (T4SS) encoded by a plasmid overproducing TraV, TraK and TraB of pED208 | | Descriptor: | TraB, TraV | | Authors: | Liu, X, Khara, P, Baker, M.L, Christie, P.J, Hu, B. | | Deposit date: | 2021-11-02 | | Release date: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structure of a type IV secretion system core complex encoded by multi-drug resistance F plasmids
Nat Commun, 13, 2022
|
|
7SPK
 
 | | Models for C16 reconstruction of Outer Membrane Core Complex (OMCC) of Type IV Secretion System (T4SS) encoded by a plasmid overproducing TraV, TraK and TraB of pED208 | | Descriptor: | TraB, TraV | | Authors: | Liu, X, Khara, P, Baker, M.L, Christie, P.J, Hu, B. | | Deposit date: | 2021-11-02 | | Release date: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of a type IV secretion system core complex encoded by multi-drug resistance F plasmids
Nat Commun, 13, 2022
|
|
7SPC
 
 | | Models for C17 reconstruction of Outer Membrane Core Complex (OMCC) of Type IV Secretion System (T4SS) encoded by F-plasmid (pED208). | | Descriptor: | TraB, TraV | | Authors: | Liu, X, Khara, P, Baker, M.L, Christie, P.J, Hu, B. | | Deposit date: | 2021-11-02 | | Release date: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure of a type IV secretion system core complex encoded by multi-drug resistance F plasmids
Nat Commun, 13, 2022
|
|
7SPB
 
 | | Models for C13 reconstruction of Outer Membrane Core Complex (OMCC) of Type IV Secretion System (T4SS) encoded by F-plasmid (pED208). | | Descriptor: | TraB, TraK, TraV | | Authors: | Liu, X, Khara, P, Baker, M.L, Christie, P.J, Hu, B. | | Deposit date: | 2021-11-02 | | Release date: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structure of a type IV secretion system core complex encoded by multi-drug resistance F plasmids
Nat Commun, 13, 2022
|
|
7SPI
 
 | | Models for C13 reconstruction of Outer Membrane Core Complex (OMCC) of Type IV Secretion System (T4SS) encoded by a plasmid overproducing TraV, TraK and TraB of pED208 | | Descriptor: | TraB, TraK, TraV | | Authors: | Liu, X, Khara, P, Baker, M.L, Christie, P.J, Hu, B. | | Deposit date: | 2021-11-02 | | Release date: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure of a type IV secretion system core complex encoded by multi-drug resistance F plasmids
Nat Commun, 13, 2022
|
|
1CF9
 
 | |
1HK7
 
 | | Middle Domain of HSP90 | | Descriptor: | CADMIUM ION, HEAT SHOCK PROTEIN HSP82, MAGNESIUM ION | | Authors: | Meyer, P, Prodromou, C, Roe, S.M, Pearl, L.H. | | Deposit date: | 2003-03-06 | | Release date: | 2004-01-29 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Analysis of the Middle Segment of Hsp90. Implications for ATP Hydrolysis and Client Protein and Cochaperone Interactions
Mol.Cell, 11, 2003
|
|
1USV
 
 | |
1USU
 
 | |
6WJ5
 
 | | Structure of human TRPA1 in complex with inhibitor GDC-0334 | | Descriptor: | (4R,5S)-4-fluoro-1-[(4-fluorophenyl)sulfonyl]-5-methyl-N-({5-(trifluoromethyl)-2-[2-(trifluoromethyl)pyrimidin-5-yl]pyridin-4-yl}methyl)-L-prolinamide, Transient receptor potential cation channel subfamily A member 1 | | Authors: | Rohou, A, Rouge, L, Arthur, C.P, Volgraf, M, Chen, H. | | Deposit date: | 2020-04-11 | | Release date: | 2021-02-17 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A TRPA1 inhibitor suppresses neurogenic inflammation and airway contraction for asthma treatment.
J.Exp.Med., 218, 2021
|
|
1QF7
 
 | |
6G4Z
 
 | | Crystal structure of murine NF-kappaB inducing kinase (NIK) in complex with compound 2f | | Descriptor: | 5-fluoranyl-1-[4-[2-[(3~{R})-1-methyl-3-oxidanyl-2-oxidanylidene-pyrrol-3-yl]ethynyl]pyridin-2-yl]indazole-3-carboxamide, Mitogen-activated protein kinase kinase kinase 14 | | Authors: | Leonardo-Silvestre, H, McEwan, P.A, Hymowitz, S.G. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Scaffold-Hopping Approach To Discover Potent, Selective, and Efficacious Inhibitors of NF-kappa B Inducing Kinase.
J. Med. Chem., 61, 2018
|
|
6G4Y
 
 | | Crystal structure of murine NF-kappaB inducing kinase (NIK) in complex with compound 1a | | Descriptor: | 10-[2-[(3~{R})-1-methyl-3-oxidanyl-2-oxidanylidene-pyrrolidin-3-yl]ethynyl]-~{N}3-(oxan-4-yl)-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepine-2,3-dicarboxamide, Mitogen-activated protein kinase kinase kinase 14, SULFATE ION | | Authors: | Hole, A.J, Hymowitz, S.G, McEwan, P.A. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Scaffold-Hopping Approach To Discover Potent, Selective, and Efficacious Inhibitors of NF-kappa B Inducing Kinase.
J. Med. Chem., 61, 2018
|
|
7R9L
 
 | | Crystal structure of HPK1 in complex with compound 2 | | Descriptor: | 2-amino-N,N-dimethyl-5-(1H-pyrrolo[2,3-b]pyridin-5-yl)benzamide, Hematopoietic progenitor kinase | | Authors: | Wu, P, Lehoux, I, Wang, W. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.332 Å) | | Cite: | Discovery of Spiro-azaindoline Inhibitors of Hematopoietic Progenitor Kinase 1 (HPK1).
Acs Med.Chem.Lett., 13, 2022
|
|
7R9N
 
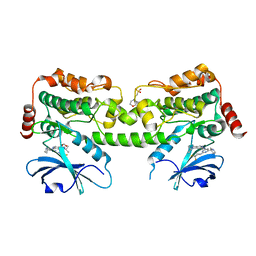 | | Crystal structure of HPK1 in complex with GNE1858 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Hematopoietic progenitor kinase, ... | | Authors: | Wu, P, Lehoux, I, Wang, W. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Spiro-azaindoline Inhibitors of Hematopoietic Progenitor Kinase 1 (HPK1).
Acs Med.Chem.Lett., 13, 2022
|
|
7R9T
 
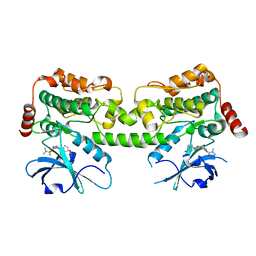 | | Crystal structure of HPK1 in complex with compound 17 | | Descriptor: | 6-amino-3-[(1S,3R)-4'-chloro-3-hydroxy-1',2'-dihydrospiro[cyclopentane-1,3'-pyrrolo[2,3-b]pyridin]-5'-yl]-2-fluoro-N,N-dimethylbenzamide, Hematopoietic progenitor kinase | | Authors: | Wu, P, Lehoux, I, Wang, W. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Spiro-azaindoline Inhibitors of Hematopoietic Progenitor Kinase 1 (HPK1).
Acs Med.Chem.Lett., 13, 2022
|
|
7R9P
 
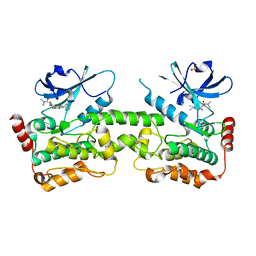 | | Crystal structure of HPK1 in complex with compound 14 | | Descriptor: | 6-amino-2-fluoro-N,N-dimethyl-3-(4'-methylspiro[cyclopropane-1,3'-pyrrolo[2,3-b]pyridin]-5'-yl)benzamide, Hematopoietic progenitor kinase, SULFATE ION | | Authors: | Wu, P, Lehoux, I, Wang, W. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery of Spiro-azaindoline Inhibitors of Hematopoietic Progenitor Kinase 1 (HPK1).
Acs Med.Chem.Lett., 13, 2022
|
|
1JK6
 
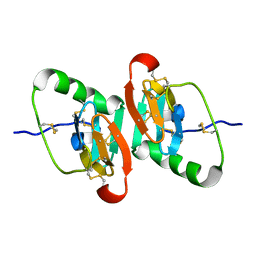 | | UNCOMPLEXED DES 1-6 BOVINE NEUROPHYSIN | | Descriptor: | NEUROPHYSIN 2 | | Authors: | Rose, J.P, Wang, B.-C. | | Deposit date: | 2001-07-11 | | Release date: | 2003-04-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of an Unliganded Neurophysin and its Vasopressin Complex: Implications for Binding and Allosteric Mechanisms
Protein Sci., 10, 2003
|
|
1JK4
 
 | | DES 1-6 BOVINE NEUROPHYSIN II COMPLEX WITH VASOPRESSIN | | Descriptor: | CADMIUM ION, Lys Vasopressin, NEUROPHYSIN 2 | | Authors: | Rose, J.P, Wang, B.-C. | | Deposit date: | 2001-07-11 | | Release date: | 2003-02-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of an Unliganded Neurophysin and its Vasopressin Complex: Implications for Binding and Allosteric Mechanisms
Protein Sci., 10, 2001
|
|
4YNE
 
 | | (R)-2-Phenylpyrrolidine Substitute Imidazopyridazines: a New Class of Potent and Selective Pan-TRK Inhibitors | | Descriptor: | 6-[(2R)-2-(3-fluorophenyl)pyrrolidin-1-yl]-3-(pyridin-2-yl)imidazo[1,2-b]pyridazine, GLYCEROL, High affinity nerve growth factor receptor, ... | | Authors: | Kreusch, A, Rucker, P, Molteni, V, Loren, J. | | Deposit date: | 2015-03-09 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.0229 Å) | | Cite: | (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
4YMJ
 
 | | (R)-2-Phenylpyrrolidine Substitute Imidazopyridazines: a New Class of Potent and Selective Pan-TRK Inhibitors | | Descriptor: | 4-[6-(benzylamino)imidazo[1,2-b]pyridazin-3-yl]benzonitrile, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kreusch, A, Rucker, P, Molteni, V, Loren, J. | | Deposit date: | 2015-03-06 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
4YPS
 
 | | (R)-2-Phenylpyrrolidine Substitute Imidazopyridazines: a New Class of Potent and Selective Pan-TRK Inhibitors | | Descriptor: | 4-{6-[(3R)-3-(3-fluorophenyl)morpholin-4-yl]imidazo[1,2-b]pyridazin-3-yl}benzonitrile, High affinity nerve growth factor receptor, SULFATE ION | | Authors: | Kreusch, A, Rucker, P, Molteni, V, Loren, J. | | Deposit date: | 2015-03-13 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1012 Å) | | Cite: | (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
6NM5
 
 | | F-pilus/MS2 Maturation protein complex | | Descriptor: | (2R)-2,3-dihydroxypropyl ethyl hydrogen (S)-phosphate, Maturation protein, Type IV conjugative transfer system pilin TraA | | Authors: | Meng, R, Chang, J, Zhang, J. | | Deposit date: | 2019-01-10 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural basis for the adsorption of a single-stranded RNA bacteriophage.
Nat Commun, 10, 2019
|
|
7OGT
 
 | | Folded elbow of cohesin | | Descriptor: | Structural maintenance of chromosomes protein 1, Structural maintenance of chromosomes protein 3 | | Authors: | Lee, B.-G, Gonzalez Llamazares, A, Collier, J, Patele, N.J, Nasmyth, K.A, Lowe, J. | | Deposit date: | 2021-05-07 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Folding of cohesin's coiled coil is important for Scc2/4-induced association with chromosomes.
Elife, 10, 2021
|
|
