1MLV
 
 | | Structure and Catalytic Mechanism of a SET Domain Protein Methyltransferase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Ribulose-1,5 biphosphate carboxylase/oxygenase large subunit N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Trievel, R.C, Beach, B.M, Dirk, L.M.A, Houtz, R.L, Hurley, J.H. | | Deposit date: | 2002-08-30 | | Release date: | 2002-10-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and catalytic mechanism of a SET domain protein methyltransferase.
Cell(Cambridge,Mass.), 111, 2002
|
|
1P0Y
 
 | | Crystal structure of the SET domain of LSMT bound to MeLysine and AdoHcy | | Descriptor: | N-METHYL-LYSINE, Ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplast, ... | | Authors: | Trievel, R.C, Flynn, E.M, Houtz, R.L, Hurley, J.H. | | Deposit date: | 2003-04-11 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Mechanism of multiple lysine methylation by the SET domain enzyme Rubisco LSMT
Nat.Struct.Biol., 10, 2003
|
|
1OZV
 
 | | Crystal structure of the SET domain of LSMT bound to Lysine and AdoHcy | | Descriptor: | LYSINE, Ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplast, ... | | Authors: | Trievel, R.C, Flynn, E.M, Houtz, R.L, Hurley, J.H. | | Deposit date: | 2003-04-09 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mechanism of multiple lysine methylation by the SET domain enzyme Rubisco LSMT
Nat.Struct.Biol., 10, 2003
|
|
3CPM
 
 | | plant peptide deformylase PDF1B crystal structure | | Descriptor: | Peptide deformylase, chloroplast, SULFATE ION, ... | | Authors: | Rodgers, D.W, Houtz, R.L, Dirk, L.M.A, Schmidt, J.J, Cai, Y. | | Deposit date: | 2008-03-31 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into the substrate specificity of plant peptide deformylase, an essential enzyme with potential for the development of novel biotechnology applications in agriculture
Biochem.J., 413, 2008
|
|
3F9W
 
 | | Structural Insights into Lysine Multiple Methylation by SET Domain Methyltransferases, SET8-Y334F / H4-Lys20 / AdoHcy | | Descriptor: | Histone H4, Histone-lysine N-methyltransferase SETD8, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Couture, J.-F, Dirk, L.M.A, Brunzelle, J.S, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2008-11-14 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural origins for the product specificity of SET domain protein methyltransferases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3F9Y
 
 | | Structural Insights into Lysine Multiple Methylation by SET Domain Methyltransferases, SET8-Y334F / H4-Lys20me1 / AdoHcy | | Descriptor: | Histone H4, Histone-lysine N-methyltransferase SETD8, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Couture, J.-F, Dirk, L.M.A, Brunzelle, J.S, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2008-11-14 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural origins for the product specificity of SET domain protein methyltransferases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3F9Z
 
 | | Structural Insights into Lysine Multiple Methylation by SET Domain Methyltransferases, SET8-Y245F / H4-Lys20 / AdoHcy | | Descriptor: | Histone H4, Histone-lysine N-methyltransferase SETD8, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Couture, J.-F, Dirk, L.M.A, Brunzelle, J.S, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2008-11-14 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural origins for the product specificity of SET domain protein methyltransferases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3F9X
 
 | | Structural Insights into Lysine Multiple Methylation by SET Domain Methyltransferases, SET8-Y334F / H4-Lys20me2 / AdoHcy | | Descriptor: | Histone H4, Histone-lysine N-methyltransferase SETD8, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Couture, J.-F, Dirk, L.M.A, Brunzelle, J.S, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2008-11-14 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural origins for the product specificity of SET domain protein methyltransferases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3M55
 
 | | SET7/9 Y305F in complex with TAF10-K189me1 peptide and AdoHcy | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, TAF10 peptide | | Authors: | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
3M53
 
 | | SET7/9 in complex with TAF10 peptide and AdoHcy | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, TAF10 peptide | | Authors: | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
3M5A
 
 | | SET7/9 Y245A in complex with TAF10-K189me3 peptide and AdoHcy | | Descriptor: | COBALT (II) ION, GLYCEROL, Histone-lysine N-methyltransferase SETD7, ... | | Authors: | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
3M58
 
 | | SET7/9 Y245A in complex with TAF10-K189me1 peptide and AdoHcy | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
3M59
 
 | | SET7/9 Y245A in complex with TAF10-K189me2 peptide and AdoHcy | | Descriptor: | COBALT (II) ION, GLYCEROL, Histone-lysine N-methyltransferase SETD7, ... | | Authors: | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
3M54
 
 | | SET7/9 Y305F in complex with TAF10 peptide and AdoHcy | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, TAF10 peptide | | Authors: | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
3M57
 
 | | SET7/9 Y245A in complex with TAF10 peptide and AdoHcy | | Descriptor: | GLYCEROL, Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
3M56
 
 | | SET7/9 Y305F in complex with TAF10-K189me2 peptide and AdoHcy | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, TAF10-K189me2 PEPTIDE | | Authors: | Del Rizzo, P.A, Couture, J.-F, Roiko, M.S, Strunk, B.S, Brunzelle, J.S, Dirk, L.M, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | SET7/9 catalytic mutants reveal the role of active site water molecules in lysine multiple methylation.
J.Biol.Chem., 285, 2010
|
|
5EG2
 
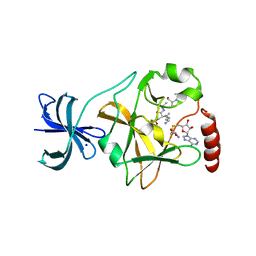 | | SET7/9 N265A in complex with AdoHcy and TAF10 peptide | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, ... | | Authors: | Kroner, G.M, Fick, R.J, Trievel, R.C. | | Deposit date: | 2015-10-26 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Sulfur-Oxygen Chalcogen Bonding Mediates AdoMet Recognition in the Lysine Methyltransferase SET7/9.
Acs Chem.Biol., 11, 2016
|
|
4J8O
 
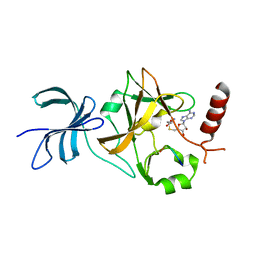 | | SET7/9 in complex with TAF10K189A peptide and AdoHcy | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, Transcription initiation factor TFIID subunit 10 | | Authors: | Horowitz, S, Trievel, R.C. | | Deposit date: | 2013-02-14 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Conservation and functional importance of carbon-oxygen hydrogen bonding in AdoMet-dependent methyltransferases.
J.Am.Chem.Soc., 135, 2013
|
|
4J7I
 
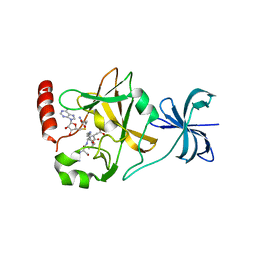 | | SET7/9Y335F in complex with TAF10 peptide and AdoHcy | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, Transcription initiation factor TFIID subunit 10 | | Authors: | Horowitz, S, Trievel, R.C. | | Deposit date: | 2013-02-13 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Conservation and functional importance of carbon-oxygen hydrogen bonding in AdoMet-dependent methyltransferases.
J.Am.Chem.Soc., 135, 2013
|
|
4J83
 
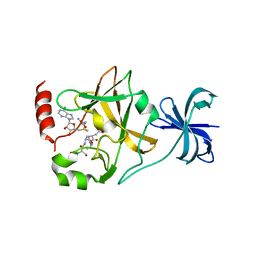 | | SET7/9 in complex with TAF10K189A peptide and AdoMet | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYLMETHIONINE, Transcription initiation factor TFIID subunit 10 | | Authors: | Horowitz, S, Nimtz, J.S, Trievel, R.C. | | Deposit date: | 2013-02-14 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conservation and functional importance of carbon-oxygen hydrogen bonding in AdoMet-dependent methyltransferases.
J.Am.Chem.Soc., 135, 2013
|
|
4J7F
 
 | |
