5X33
 
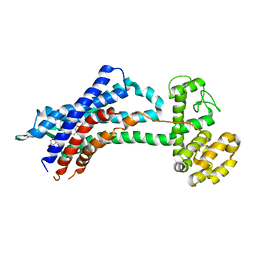 | | Leukotriene B4 receptor BLT1 in complex with BIIL260 | | Descriptor: | 4-[[3-[[4-[2-(4-hydroxyphenyl)propan-2-yl]phenoxy]methyl]phenyl]methoxy]benzenecarboximidamide, LTB4 receptor,Lysozyme,LTB4 receptor | | Authors: | Hori, T, Hirata, K, Yamashita, K, Kawano, Y, Yamamoto, M, Yokoyama, S. | | Deposit date: | 2017-02-03 | | Release date: | 2018-01-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Na+-mimicking ligands stabilize the inactive state of leukotriene B4receptor BLT1.
Nat. Chem. Biol., 14, 2018
|
|
1GE6
 
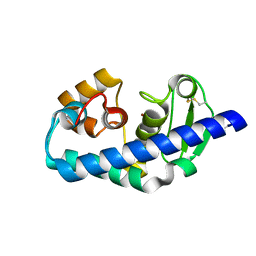 | | ZINC PEPTIDASE FROM GRIFOLA FRONDOSA | | Descriptor: | PEPTIDYL-LYS METALLOENDOPEPTIDASE, ZINC ION, alpha-D-mannopyranose | | Authors: | Hori, T, Kumasaka, T, Yamamoto, M, Nonaka, T, Tanaka, N, Hashimoto, Y, Ueki, T, Takio, K. | | Deposit date: | 2000-10-11 | | Release date: | 2001-03-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a new 'aspzincin' metalloendopeptidase from Grifola frondosa: implications for the catalytic mechanism and substrate specificity based on several different crystal forms.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GE5
 
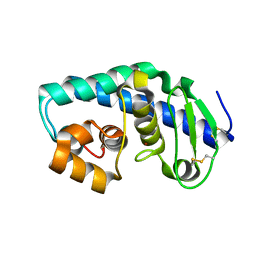 | | ZINC PEPTIDASE FROM GRIFOLA FRONDOSA | | Descriptor: | PEPTIDYL-LYS METALLOENDOPEPTIDASE, ZINC ION, alpha-D-mannopyranose | | Authors: | Hori, T, Kumasaka, T, Yamamoto, M, Nonaka, T, Tanaka, N, Hashimoto, Y, Ueki, T, Takio, K. | | Deposit date: | 2000-10-11 | | Release date: | 2001-03-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a new 'aspzincin' metalloendopeptidase from Grifola frondosa: implications for the catalytic mechanism and substrate specificity based on several different crystal forms.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GE7
 
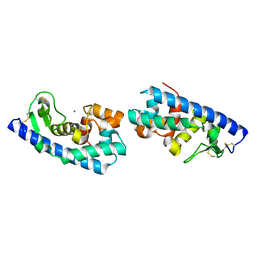 | | ZINC PEPTIDASE FROM GRIFOLA FRONDOSA | | Descriptor: | PEPTIDYL-LYS METALLOENDOPEPTIDASE, ZINC ION, alpha-D-mannopyranose | | Authors: | Hori, T, Kumasaka, T, Yamamoto, M, Nonaka, T, Tanaka, N, Hashimoto, Y, Ueki, T, Takio, K. | | Deposit date: | 2000-10-11 | | Release date: | 2001-03-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a new 'aspzincin' metalloendopeptidase from Grifola frondosa: implications for the catalytic mechanism and substrate specificity based on several different crystal forms.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1G12
 
 | | ZINC PEPTIDASE FROM GRIFOLA FRONDOSA | | Descriptor: | PEPTIDYL-LYS METALLOENDOPEPTIDASE, ZINC ION, alpha-D-mannopyranose | | Authors: | Hori, T, Kumasaka, T, Yamamoto, M, Nonaka, T, Tanaka, N, Hashimoto, Y, Ueki, T, Takio, K. | | Deposit date: | 2000-10-10 | | Release date: | 2001-03-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a new 'aspzincin' metalloendopeptidase from Grifola frondosa: implications for the catalytic mechanism and substrate specificity based on several different crystal forms.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2DM6
 
 | | Crystal structure of anti-configuration of indomethacin and leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase complex | | Descriptor: | INDOMETHACIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent leukotriene B4 12-hydroxydehydrogenase, ... | | Authors: | Hori, T, Ishijima, J, Yokomizo, T, Ago, H, Shimizu, T, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-20 | | Release date: | 2006-11-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of anti-configuration of indomethacin and leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase complex reveals the structural basis of broad spectrum indomethacin efficacy
J.Biochem.(Tokyo), 140, 2006
|
|
1V3V
 
 | | Crystal structure of leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase complexed with NADP and 15-oxo-PGE2 | | Descriptor: | (5E,13E)-11-HYDROXY-9,15-DIOXOPROSTA-5,13-DIEN-1-OIC ACID, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hori, T, Yokomizo, T, Ago, H, Sugahara, M, Ueno, G, Yamamoto, M, Kumasaka, T, Shimizu, T, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-06 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of leukotriene B4 12-hydroxydehydrogenase/15-Oxo-prostaglandin 13-reductase catalytic mechanism and a possible Src homology 3 domain binding loop
J.Biol.Chem., 279, 2004
|
|
1V3T
 
 | | Crystal structure of leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, leukotriene b4 12-hydroxydehydrogenase/prostaglandin 15-keto reductase | | Authors: | Hori, T, Yokomizo, T, Ago, H, Sugahara, M, Ueno, G, Yamamoto, M, Kumasaka, T, Shimizu, T, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-05 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of leukotriene B4 12-hydroxydehydrogenase/15-Oxo-prostaglandin 13-reductase catalytic mechanism and a possible Src homology 3 domain binding loop
J.Biol.Chem., 279, 2004
|
|
1V3U
 
 | | Crystal structure of leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase in apo form | | Descriptor: | CHLORIDE ION, leukotriene b4 12-hydroxydehydrogenase/prostaglandin 15-keto reductase | | Authors: | Hori, T, Yokomizo, T, Ago, H, Sugahara, M, Ueno, G, Yamamoto, M, Kumasaka, T, Shimizu, T, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-05 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of leukotriene B4 12-hydroxydehydrogenase/15-Oxo-prostaglandin 13-reductase catalytic mechanism and a possible Src homology 3 domain binding loop
J.Biol.Chem., 279, 2004
|
|
3B0D
 
 | | Crystal structure of the chicken CENP-T histone fold/CENP-W complex, crystal form II | | Descriptor: | CITRIC ACID, Centromere protein T, Centromere protein W | | Authors: | Nishino, T, Takeuchi, K, Gascoigne, K.E, Suzuki, A, Hori, T, Oyama, T, Morikawa, K, Cheeseman, I.M, Fukagawa, T. | | Deposit date: | 2011-06-08 | | Release date: | 2012-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | CENP-T-W-S-X Forms a Unique Centromeric Chromatin Structure with a Histone-like Fold.
Cell(Cambridge,Mass.), 148, 2012
|
|
3B0C
 
 | | Crystal structure of the chicken CENP-T histone fold/CENP-W complex, crystal form I | | Descriptor: | CITRIC ACID, Centromere protein T, Centromere protein W | | Authors: | Nishino, T, Takeuchi, K, Gascoigne, K.E, Suzuki, A, Hori, T, Oyama, T, Morikawa, K, Cheeseman, I.M, Fukagawa, T. | | Deposit date: | 2011-06-08 | | Release date: | 2012-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | CENP-T-W-S-X Forms a Unique Centromeric Chromatin Structure with a Histone-like Fold.
Cell(Cambridge,Mass.), 148, 2012
|
|
3B0B
 
 | | Crystal structure of the chicken CENP-S/CENP-X complex | | Descriptor: | Centromere protein S, Centromere protein X | | Authors: | Nishino, T, Takeuchi, K, Gascoigne, K.E, Suzuki, A, Hori, T, Oyama, T, Morikawa, K, Cheeseman, I.M, Fukagawa, T. | | Deposit date: | 2011-06-08 | | Release date: | 2012-03-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | CENP-T-W-S-X Forms a Unique Centromeric Chromatin Structure with a Histone-like Fold.
Cell(Cambridge,Mass.), 148, 2012
|
|
3VH6
 
 | | Crystal structure of the chicken CENP-T histone fold/CENP-W/CENP-S/CENP-X heterotetrameric complex, crystal form II | | Descriptor: | CENP-S, CENP-T, CENP-W, ... | | Authors: | Nishino, T, Takeuchi, K, Gascoigne, K.E, Suzuki, A, Hori, T, Oyama, T, Morikawa, K, Cheeseman, I.M, Fukagawa, T. | | Deposit date: | 2011-08-23 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.351 Å) | | Cite: | CENP-T-W-S-X Forms a Unique Centromeric Chromatin Structure with a Histone-like Fold
Cell(Cambridge,Mass.), 148, 2012
|
|
3VH5
 
 | | Crystal structure of the chicken CENP-T histone fold/CENP-W/CENP-S/CENP-X heterotetrameric complex, crystal form I | | Descriptor: | CENP-S, CENP-T, CENP-W, ... | | Authors: | Nishino, T, Takeuchi, K, Gascoigne, K.E, Suzuki, A, Hori, T, Oyama, T, Morikawa, K, Cheeseman, I.M, Fukagawa, T. | | Deposit date: | 2011-08-23 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | CENP-T-W-S-X Forms a Unique Centromeric Chromatin Structure with a Histone-like Fold
Cell(Cambridge,Mass.), 148, 2012
|
|
1V26
 
 | | Crystal structure of tt0168 from Thermus thermophilus HB8 | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, MYRISTIC ACID, ... | | Authors: | Hisanaga, Y, Ago, H, Nakatsu, T, Hamada, K, Ida, K, Kanda, H, Yamamoto, M, Hori, T, Arii, Y, Sugahara, M, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-07 | | Release date: | 2004-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of the Substrate-specific Two-step Catalysis of Long Chain Fatty Acyl-CoA Synthetase Dimer
J.Biol.Chem., 279, 2004
|
|
1ULT
 
 | | Crystal structure of tt0168 from Thermus thermophilus HB8 | | Descriptor: | CITRIC ACID, long chain fatty acid-CoA ligase | | Authors: | Hisanaga, Y, Ago, H, Nakatsu, T, Hamada, K, Ida, K, Kanda, H, Yamamoto, M, Hori, T, Arii, Y, Sugahara, M, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-16 | | Release date: | 2004-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of the substrate specific two-step catalysis of long chain fatty acyl-CoA synthetase dimer
J.Biol.Chem., 279, 2004
|
|
1V25
 
 | | Crystal structure of tt0168 from Thermus thermophilus HB8 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, long-chain-fatty-acid-CoA synthetase | | Authors: | Hisanaga, Y, Ago, H, Nakatsu, T, Hamada, K, Ida, K, Kanda, H, Yamamoto, M, Hori, T, Arii, Y, Sugahara, M, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-07 | | Release date: | 2004-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Substrate-specific Two-step Catalysis of Long Chain Fatty Acyl-CoA Synthetase Dimer
J.Biol.Chem., 279, 2004
|
|
1F88
 
 | | CRYSTAL STRUCTURE OF BOVINE RHODOPSIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MERCURY (II) ION, RETINAL, ... | | Authors: | Okada, T, Palczewski, K, Stenkamp, R.E, Miyano, M. | | Deposit date: | 2000-06-29 | | Release date: | 2000-08-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of rhodopsin: A G protein-coupled receptor.
Science, 289, 2000
|
|
2ZIY
 
 | | Crystal structure of squid rhodopsin | | Descriptor: | PALMITIC ACID, RETINAL, Rhodopsin | | Authors: | Miyano, M, Shimamura, T. | | Deposit date: | 2008-02-27 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of squid rhodopsin with intracellularly extended cytoplasmic region
J.Biol.Chem., 283, 2008
|
|
5ZWI
 
 | | Interaction between Vitamin D receptor (VDR) and a ligand having a dienone group | | Descriptor: | (2S)-2-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]oct-4,6-diene-3-one, 13-meric peptide from DRIP205 NR2 BOX peptide, Vitamin D3 receptor | | Authors: | Yoshizawa, M, Itoh, T, Anami, Y, Kato, A, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-05-15 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of the Histidine Residue in Vitamin D Receptor That Covalently Binds to Electrophilic Ligands
J. Med. Chem., 61, 2018
|
|
5ZWH
 
 | | Covalent bond formation between histidine of Vitamin D receptor (VDR) and a full agonist having an ene-ynone group via conjugate addition reaction | | Descriptor: | (2S)-2-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]oct-4,6-diene-3-one, (E,2S)-2-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]oct-6-en-4-yn-3-one, 13-meric peptide from DRIP205 NR2 BOX peptide, ... | | Authors: | Yoshizawa, M, Itoh, T, Anami, Y, Kato, A, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-05-15 | | Release date: | 2018-07-18 | | Last modified: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Identification of the Histidine Residue in Vitamin D Receptor That Covalently Binds to Electrophilic Ligands
J. Med. Chem., 61, 2018
|
|
5ZWF
 
 | | Covalent bond formation between histidine of Vitamin D receptor (VDR) and a full agonist having a enone with a beta methyl group via conjugate addition reaction | | Descriptor: | (E,7R)-7-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]oct-2-en-4-one, 13-meric peptide from DRIP205 NR2 BOX peptide, Vitamin D3 receptor | | Authors: | Yoshizawa, M, Itoh, T, Anami, Y, Kato, A, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-05-15 | | Release date: | 2018-07-18 | | Last modified: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of the Histidine Residue in Vitamin D Receptor That Covalently Binds to Electrophilic Ligands
J. Med. Chem., 61, 2018
|
|
5ZWE
 
 | | Covalent bond formation between histidine of Vitamin D receptor (VDR) and a full agonist having a vinyl ketone group via conjugate addition reaction | | Descriptor: | (6R)-6-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]hept-1-en-3-one, 13-meric peptide from DRIP205 NR2 BOX peptide, Vitamin D3 receptor | | Authors: | Yoshizawa, M, Itoh, T, Anami, Y, Kato, A, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-05-15 | | Release date: | 2018-07-18 | | Last modified: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Identification of the Histidine Residue in Vitamin D Receptor That Covalently Binds to Electrophilic Ligands
J. Med. Chem., 61, 2018
|
|
5Z23
 
 | | Crystal structure of the nucleosome containing a chimeric histone H3/CENP-A CATD | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Arimura, Y, Tachiwana, H, Takagi, H. | | Deposit date: | 2017-12-28 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The CENP-A centromere targeting domain facilitates H4K20 monomethylation in the nucleosome by structural polymorphism.
Nat Commun, 10, 2019
|
|
5ZBX
 
 | | The crystal structure of the nucleosome containing histone H3.1 CATD(V76Q, K77D) | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Arimura, Y, Takagi, H, Kurumizaka, H. | | Deposit date: | 2018-02-13 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The CENP-A centromere targeting domain facilitates H4K20 monomethylation in the nucleosome by structural polymorphism.
Nat Commun, 10, 2019
|
|
