3PYY
 
 | | Discovery and Characterization of a Cell-Permeable, Small-molecule c-Abl Kinase Activator that Binds to the Myristoyl Binding Site | | Descriptor: | (5R)-5-[3-(4-fluorophenyl)-1-phenyl-1H-pyrazol-4-yl]imidazolidine-2,4-dione, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, GLYCEROL, ... | | Authors: | Yang, J, Campobasso, N, Biju, M.P, Fisher, K, Pan, X.Q, Cottom, J, Galbraith, S, Ho, T, Zhang, H, Hong, X, Ward, P, Hofmann, G, Siegfried, B. | | Deposit date: | 2010-12-13 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and Characterization of a Cell-Permeable, Small-Molecule c-Abl Kinase Activator that Binds to the Myristoyl Binding Site.
Chem.Biol., 18, 2011
|
|
1T15
 
 | | Crystal Structure of the Brca1 BRCT Domains in Complex with the Phosphorylated Interacting Region from Bach1 Helicase | | Descriptor: | BRCA1 interacting protein C-terminal helicase 1, Breast cancer type 1 susceptibility protein | | Authors: | Clapperton, J.A, Manke, I.A, Lowery, D.M, Ho, T, Haire, L.F, Yaffe, M.B, Smerdon, S.J. | | Deposit date: | 2004-04-15 | | Release date: | 2004-05-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer
Nat.Struct.Mol.Biol., 11, 2004
|
|
6MX3
 
 | | Crystal structure of human STING (G230A, H232R, R293Q) in complex with Compound 1 | | Descriptor: | (3S,4S)-2-(4-tert-butylphenyl)-3-(4-methoxyphenyl)-1-oxo-1,2,3,4-tetrahydroisoquinoline-4-carboxylic acid, CALCIUM ION, Stimulator of interferon genes protein | | Authors: | Lesburg, C.A, Siu, T, Ho, T. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.362 Å) | | Cite: | Discovery of a Novel cGAMP Competitive Ligand of the Inactive Form of STING.
ACS Med Chem Lett, 10, 2019
|
|
6MXE
 
 | | Crystal structure of human STING (G230A, H232R, R293Q) in complex with Compound 18 | | Descriptor: | CALCIUM ION, Stimulator of interferon genes protein, [(3S,4S)-2-(4-tert-butyl-3-chlorophenyl)-3-(2,3-dihydro-1,4-benzodioxin-6-yl)-7-fluoro-1-oxo-1,2,3,4-tetrahydroisoquinolin-4-yl]acetic acid | | Authors: | Lesburg, C.A, Siu, T, Ho, T. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Discovery of a Novel cGAMP Competitive Ligand of the Inactive Form of STING.
ACS Med Chem Lett, 10, 2019
|
|
6MX0
 
 | | Crystal structure of human STING apoprotein (G230A, H232R, R293Q) | | Descriptor: | CALCIUM ION, Stimulator of interferon genes protein | | Authors: | Lesburg, C.A, Siu, T, Ho, T. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of a Novel cGAMP Competitive Ligand of the Inactive Form of STING.
ACS Med Chem Lett, 10, 2019
|
|
6X9I
 
 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*AP*GP*GP*CP*(5CM)P*GP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*G)-R(P*(PYO))-D(P*GP*GP*CP*CP*TP*C)-3'), ... | | Authors: | Pathuri, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-06-02 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia.
Nat Cancer, 2, 2021
|
|
6X9K
 
 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Inhibitor GSK3685032A | | Descriptor: | (2R)-2-{[6-(4-aminopiperidin-1-yl)-3,5-dicyano-4-ethylpyridin-2-yl]sulfanyl}-2-phenylacetamide, 1,2-ETHANEDIOL, DNA (5'-D(*GP*AP*GP*GP*CP*(5CM)P*GP*CP*CP*TP*GP*C)-3'), ... | | Authors: | Pathuri, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-06-02 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia.
Nat Cancer, 2, 2021
|
|
6X9J
 
 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Inhibitor GSK3830052 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*AP*GP*GP*CP*(5CM)P*GP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*G)-R(P*(PYO))-D(P*GP*GP*CP*CP*TP*C)-3'), ... | | Authors: | Pathuri, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-06-02 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia.
Nat Cancer, 2, 2021
|
|
6X3L
 
 | | Sortilin-Progranulin Interaction With Compound 2 | | Descriptor: | 1-benzyl-3-tert-butyl-1H-pyrazole-5-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Parthasarathy, G, Soisson, S.M. | | Deposit date: | 2020-05-21 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of potent inhibitors of the sortilin-progranulin interaction.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6X48
 
 | | Sortilin-Progranulin Interaction With Compound 17 | | Descriptor: | GLYCEROL, N-(3,5-dichlorobenzene-1-carbonyl)-5,5-dimethyl-L-norleucine, Sortilin, ... | | Authors: | Parthasarathy, G, Soisson, S.M. | | Deposit date: | 2020-05-22 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of potent inhibitors of the sortilin-progranulin interaction.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6X4H
 
 | | Sortilin-Progranulin Interaction With Compound 24 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-methyl-N-(6-phenoxypyridine-3-carbonyl)-L-leucine, GLYCEROL, ... | | Authors: | Parthasarathy, G, Soisson, S.M, Klein, D. | | Deposit date: | 2020-05-22 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of potent inhibitors of the sortilin-progranulin interaction.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
3PXE
 
 | |
3PXB
 
 | |
3PXC
 
 | |
3PXD
 
 | |
3PXA
 
 | |
5TZY
 
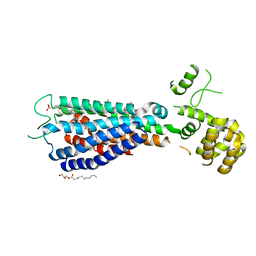 | | GPR40 in complex with AgoPAM AP8 and partial agonist MK-8666 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,3R)-3-cyclopropyl-3-[(2R)-2-(1-{(1S)-1-[5-fluoro-2-(trifluoromethoxy)phenyl]ethyl}piperidin-4-yl)-3,4-dihydro-2H-1-benzopyran-7-yl]-2-methylpropanoic acid, (5aR,6S,6aS)-3-({2',6'-dimethyl-4'-[3-(methylsulfonyl)propoxy][1,1'-biphenyl]-3-yl}methoxy)-5,5a,6,6a-tetrahydrocyclopropa[4,5]cyclopenta[1,2-c]pyridine-6-carboxylic acid, ... | | Authors: | Lu, J, Byrne, N, Patel, S, Sharma, S, Soisson, S.M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5TZR
 
 | | GPR40 in complex with partial agonist MK-8666 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (5aR,6S,6aS)-3-({2',6'-dimethyl-4'-[3-(methylsulfonyl)propoxy][1,1'-biphenyl]-3-yl}methoxy)-5,5a,6,6a-tetrahydrocyclopropa[4,5]cyclopenta[1,2-c]pyridine-6-carboxylic acid, Free fatty acid receptor 1,Endolysin,Free fatty acid receptor 1, ... | | Authors: | Lu, J, Byrne, N, Patel, S, Sharma, S, Soisson, S.M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6NPE
 
 | | C-abl Kinase domain with the activator(cmpd6), 2-cyano-N-(4-(3,4-dichlorophenyl)thiazol-2-yl)acetamide | | Descriptor: | 2-cyano-~{N}-[4-(3,4-dichlorophenyl)-1,3-thiazol-2-yl]ethanamide, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, NONAETHYLENE GLYCOL, ... | | Authors: | campobasso, N. | | Deposit date: | 2019-01-17 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification and Optimization of Novel Small c-Abl Kinase Activators Using Fragment and HTS Methodologies.
J. Med. Chem., 62, 2019
|
|
6NPV
 
 | |
6NPU
 
 | |
4YKN
 
 | | Pi3K alpha lipid kinase with Active Site Inhibitor | | Descriptor: | 3-(6-methoxypyridin-3-yl)-5-[({4-[(5-methyl-1,3,4-thiadiazol-2-yl)sulfamoyl]phenyl}amino)methyl]benzoic acid, Phosphatidylinositol 3-kinase regulatory subunit alpha,Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform fusion protein | | Authors: | Elkins, P.A. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of a Potent Class of PI3K alpha Inhibitors with Unique Binding Mode via Encoded Library Technology (ELT).
Acs Med.Chem.Lett., 6, 2015
|
|
