3M9I
 
 | |
5U76
 
 | |
5U70
 
 | |
5A6E
 
 | | Cryo-EM structure of the Slo2.2 Na-activated K channel | | Descriptor: | GATING RING OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, PORE DOMAIN OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, RCK2 ELABORATION OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, ... | | Authors: | Hite, R.K, Yuan, P, Li, Z, Hsuing, Y, Walz, T, MacKinnon, R. | | Deposit date: | 2015-06-25 | | Release date: | 2015-10-14 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-Electron Microscopy Structure of the Slo2.2 Na1-Activated K1 Channel
Nature, 527, 2015
|
|
5A6G
 
 | | Cryo-EM structure of the Slo2.2 Na-activated K channel | | Descriptor: | PORE DOMAIN OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, S1-S4 DOMAIN OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1 | | Authors: | Hite, R.K, Yuan, P, Li, Z, Hsuing, Y, Walz, T, MacKinnon, R. | | Deposit date: | 2015-06-25 | | Release date: | 2015-10-14 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-Electron Microscopy Structure of the Slo2.2 Na1-Activated K1 Channel
Nature, 527, 2015
|
|
5A6F
 
 | | Cryo-EM structure of the Slo2.2 Na-activated K channel | | Descriptor: | GATING RING OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1, RCK2 ELABORATION OF POTASSIUM CHANNEL SUBFAMILY T MEMBER 1 | | Authors: | Hite, R.K, Yuan, P, Li, Z, Hsuing, Y, Walz, T, MacKinnon, R. | | Deposit date: | 2015-06-25 | | Release date: | 2015-10-14 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-Electron Microscopy Structure of the Slo2.2 Na1-Activated K1 Channel
Nature, 527, 2015
|
|
8UBX
 
 | | Ethanolamine-bound FLVCR1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, ETHANOLAMINE, Heme transporter FLVCR1 | | Authors: | Hite, R.K, Son, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1
To Be Published
|
|
8UC0
 
 | | Endogenous ligand bound FLVCR1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Heme transporter FLVCR1 | | Authors: | Hite, R.K, Son, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1
To Be Published
|
|
8UBW
 
 | | Choline-bound FLVCR1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, CHOLINE ION, Heme transporter FLVCR1 | | Authors: | Hite, R.K, Son, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1
To Be Published
|
|
8UBZ
 
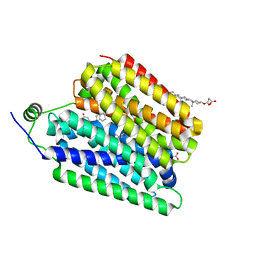 | | Choline-bound FLVCR1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, CHOLINE ION, Heme transporter FLVCR1 | | Authors: | Hite, R.K, Son, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1
To Be Published
|
|
8UBY
 
 | | Choline-bound FLVCR1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, CHOLINE ION, Heme transporter FLVCR1 | | Authors: | Hite, R.K, Son, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1
To Be Published
|
|
5TJI
 
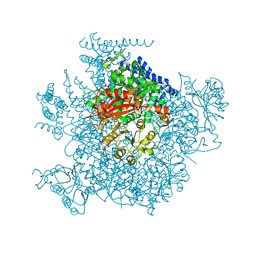 | | Ca2+ bound aplysia Slo1 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, High conductance calcium-activated potassium channel | | Authors: | MacKinnon, R, Tao, X, Hite, R.K. | | Deposit date: | 2016-10-04 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for gating the high-conductance Ca(2+)-activated K(+) channel.
Nature, 541, 2017
|
|
6DQV
 
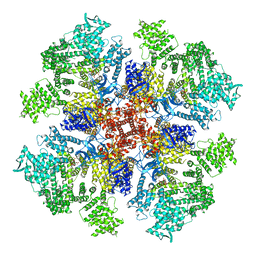 | | Class 2 IP3-bound human type 3 1,4,5-inositol trisphosphate receptor | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DQN
 
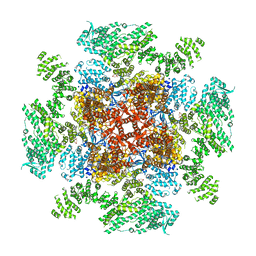 | | Class 1 IP3-bound human type 3 1,4,5-inositol trisphosphate receptor | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DR0
 
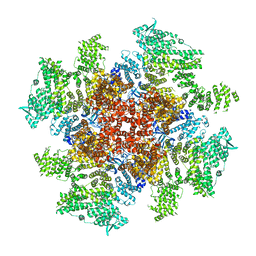 | | Class 5 IP3-bound human type 3 1,4,5-inositol trisphosphate receptor | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2018-08-15 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DRC
 
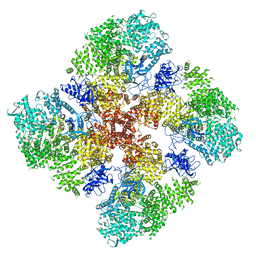 | | High IP3 Ca2+ human type 3 1,4,5-inositol trisphosphate receptor | | Descriptor: | CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ... | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DQZ
 
 | | Class 4 IP3-bound human type 3 1,4,5-inositol trisphosphate receptor | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (6.01 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DQS
 
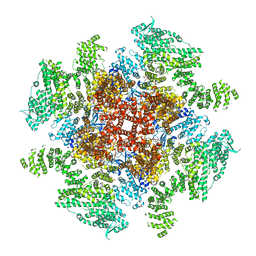 | | Class 3 IP3-bound human type 3 1,4,5-inositol trisphosphate receptor | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DRA
 
 | | Low IP3 Ca2+ human type 3 1,4,5-inositol trisphosphate receptor | | Descriptor: | CALCIUM ION, Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DQJ
 
 | | Human type 3 1,4,5-inositol trisphosphate receptor in a ligand-free state | | Descriptor: | Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2018-08-15 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DR2
 
 | | Ca2+-bound human type 3 1,4,5-inositol trisphosphate receptor | | Descriptor: | CALCIUM ION, Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-07-18 | | Last modified: | 2018-08-15 | | Method: | ELECTRON MICROSCOPY (4.33 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5TJ6
 
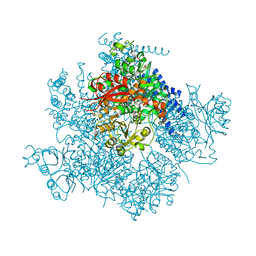 | | Ca2+ bound aplysia Slo1 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CALCIUM ION, High conductance calcium-activated potassium channel, ... | | Authors: | MacKinnon, R, Tao, X, Hite, R.K. | | Deposit date: | 2016-10-03 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the open high-conductance Ca(2+)-activated K(+) channel.
Nature, 541, 2017
|
|
7JJO
 
 | | Structural Basis of the Activation of Heterotrimeric Gs-protein by Isoproterenol-bound Beta1-Adrenergic Receptor | | Descriptor: | Beta1-Adrenergic Receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Su, M, Zhu, L, Zhang, Y, Paknejad, N, Dey, R, Huang, J, Lee, M.Y, Williams, D, Jordan, K.D, Eng, E.T, Ernst, O.P, Meyerson, J.R, Hite, R.K, Walz, T, Liu, W, Huang, X.Y. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural Basis of the Activation of Heterotrimeric Gs-Protein by Isoproterenol-Bound beta 1 -Adrenergic Receptor.
Mol.Cell, 80, 2020
|
|
6NZZ
 
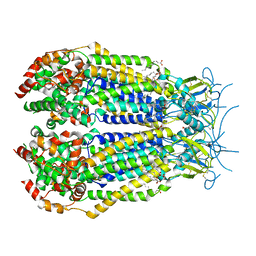 | | LRRC8A-DCPIB in MSP1E3D1 nanodisc expanded state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 4-{[(2S)-2-butyl-6,7-dichloro-2-cyclopentyl-1-oxo-2,3-dihydro-1H-inden-5-yl]oxy}butanoic acid, Volume-regulated anion channel subunit LRRC8A | | Authors: | Kern, D.M, Hite, R.K, Brohawn, S.G. | | Deposit date: | 2019-02-14 | | Release date: | 2019-02-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-EM structures of the DCPIB-inhibited volume-regulated anion channel LRRC8A in lipid nanodiscs.
Elife, 8, 2019
|
|
6O00
 
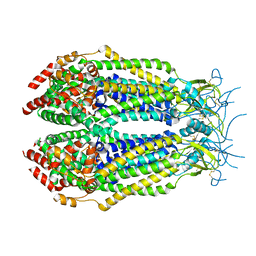 | |
