1IQ6
 
 | | (R)-HYDRATASE FROM A. CAVIAE INVOLVED IN PHA BIOSYNTHESIS | | Descriptor: | (R)-SPECIFIC ENOYL-COA HYDRATASE, ISOPROPYL ALCOHOL | | Authors: | Hisano, T, Tsuge, T, Fukui, T, Iwata, T, Doi, Y. | | Deposit date: | 2001-06-18 | | Release date: | 2003-02-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the (R)-specific enoyl-CoA hydratase from Aeromonas caviae involved in polyhydroxyalkanoate
biosynthesis
J.Biol.Chem., 278, 2003
|
|
1JUD
 
 | | L-2-HALOACID DEHALOGENASE | | Descriptor: | L-2-HALOACID DEHALOGENASE | | Authors: | Hisano, T, Hata, Y, Fujii, T, Liu, J.-Q, Kurihara, T, Esaki, N, Soda, K. | | Deposit date: | 1996-06-03 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of L-2-haloacid dehalogenase from Pseudomonas sp. YL. An alpha/beta hydrolase structure that is different from the alpha/beta hydrolase fold.
J.Biol.Chem., 271, 1996
|
|
2D80
 
 | | Crystal structure of PHB depolymerase from Penicillium funiculosum | | Descriptor: | PHB depolymerase, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Hisano, T, Kasuya, K, Saito, T, Iwata, T, Miki, K. | | Deposit date: | 2005-11-30 | | Release date: | 2006-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Polyhydroxybutyrate Depolymerase from Penicillium funiculosum Provides Insights into the Recognition and Degradation of Biopolyesters
J.Mol.Biol., 356, 2006
|
|
2D81
 
 | | PHB depolymerase (S39A) complexed with R3HB trimer | | Descriptor: | (1R)-3-{[(1R)-3-METHOXY-1-METHYL-3-OXOPROPYL]OXY}-1-METHYL-3-OXOPROPYL (3R)-3-HYDROXYBUTANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, PHB depolymerase | | Authors: | Hisano, T, Kasuya, K, Saito, T, Iwata, T, Miki, K. | | Deposit date: | 2005-11-30 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The Crystal Structure of Polyhydroxybutyrate Depolymerase from Penicillium funiculosum Provides Insights into the Recognition and Degradation of Biopolyesters
J.Mol.Biol., 356, 2006
|
|
7W78
 
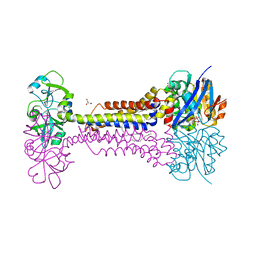 | | Heme exporter HrtBA in complex with Mg-AMPPNP | | Descriptor: | ACETATE ION, DODECANE, GLYCEROL, ... | | Authors: | Hisano, T, Nakamura, H, Rahman, M.M, Tosha, T, Shirouzu, M, Shiro, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.884 Å) | | Cite: | Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7W79
 
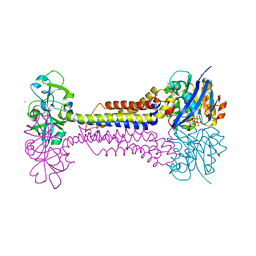 | | Heme exporter HrtBA in complex with Mn-AMPPNP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, Putative ABC transport system integral membrane protein, ... | | Authors: | Hisano, T, Nakamura, H, Rahman, M.M, Tosha, T, Shirouzu, M, Shiro, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-06-22 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7W7B
 
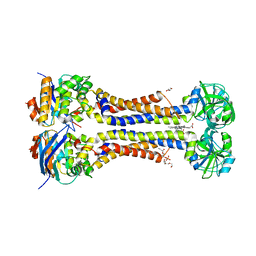 | | Heme exporter HrtBA in complex with protoporphyrin IX containing manganese(III), high resolution data | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, DI(HYDROXYETHYL)ETHER, PROTOPORPHYRIN IX CONTAINING MN, ... | | Authors: | Hisano, T, Nakamura, H, Rahman, M.M, Tosha, T, Shirouzu, M, Shiro, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7W7A
 
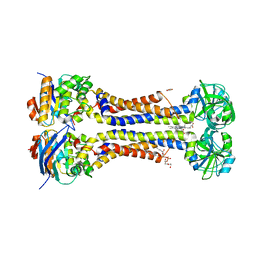 | | Heme exporter in complex with Mn-containing protoporphyrin IX, Mn-anomalous data | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hisano, T, Nakamura, H, Rahman, M.M, Tosha, T, Shirouzu, M, Shiro, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7W7D
 
 | | Heme exporter HrtBA in complex with heme | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative ABC transport system integral membrane protein, Putative ABC transport system, ... | | Authors: | Hisano, T, Nakamura, H, Rahman, M.M, Tosha, T, Shiro, Y, Shirouzu, M. | | Deposit date: | 2021-12-04 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6FWF
 
 | | Low resolution structure of Neisseria meningitidis qNOR | | Descriptor: | CALCIUM ION, FE (III) ION, Nitric-oxide reductase, ... | | Authors: | Young, D, Antonyuk, S, Tosha, T, Hisano, T, Hasnain, S, Shiro, Y. | | Deposit date: | 2018-03-06 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Characterization of the quinol-dependent nitric oxide reductase from the pathogen Neisseria meningitidis, an electrogenic enzyme.
Sci Rep, 8, 2018
|
|
3HRX
 
 | |
5Y5K
 
 | | Time-resolved SFX structure of cytochrome P450nor : 20 ms after photo-irradiation of caged NO in the absence of NADH (NO-bound state), light data | | Descriptor: | NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5CPG
 
 | | R-Hydratase PhaJ1 from Pseudomonas aeruginosa in the unliganded form | | Descriptor: | (R)-specific enoyl-CoA hydratase, GLYCEROL | | Authors: | Tsuge, T, Sato, S, Hiroe, A, Ishizuka, K, Kanazawa, H, Kanagarajan, S, Shiro, Y, Hisano, T. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Contribution of the Distal Pocket Residue to the Acyl-Chain-Length Specificity of (R)-Specific Enoyl-Coenzyme A Hydratases from Pseudomonas spp.
Appl.Environ.Microbiol., 81, 2015
|
|
1ZRN
 
 | | INTERMEDIATE STRUCTURE OF L-2-HALOACID DEHALOGENASE WITH MONOCHLOROACETATE | | Descriptor: | ACETIC ACID, L-2-HALOACID DEHALOGENASE | | Authors: | Li, Y.-F, Hata, Y, Fujii, T, Hisano, T, Nishihara, M, Kurihara, T, Esaki, N. | | Deposit date: | 1998-03-03 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of reaction intermediates of L-2-haloacid dehalogenase and implications for the reaction mechanism.
J.Biol.Chem., 273, 1998
|
|
7DVO
 
 | | Structure of Reaction Intermediate of Cytochrome P450 NO Reductase (P450nor) Determined by XFEL | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Nomura, T, Kimura, T, Kanematsu, Y, Yamashita, K, Hirata, K, Ueno, G, Murakami, H, Hisano, T, Yamagiwa, R, Takeda, H, Gopalasingam, C, Yuki, K, Kousaka, R, Yanagasawa, S, Shoji, O, Kumasaka, T, Takano, Y, Ago, H, Yamamoto, M, Sugimoto, H, Tosha, T, Kubo, M, Shiro, Y. | | Deposit date: | 2021-01-14 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Short-lived intermediate in N 2 O generation by P450 NO reductase captured by time-resolved IR spectroscopy and XFEL crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5Y5M
 
 | | SFX structure of cytochrome P450nor: a complete dark data without pump laser (resting state) | | Descriptor: | NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5L
 
 | | Time-resolved SFX structure of cytochrome P450nor: dark-2 data in the absence of NADH (resting state) | | Descriptor: | NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5I
 
 | | Time-resolved SFX structure of cytochrome P450nor: 20 ms after photo-irradiation of caged NO in the presence of NADH (NO-bound state), light data | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5J
 
 | | Time-resolved SFX structure of cytochrome P450nor: dark-2 data in the presence of NADH (resting state) | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5H
 
 | | SF-ROX structure of cytochrome P450nor (NO-bound state) determined at SACLA | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Yamagiwa, R, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Yamashita, A, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5XSO
 
 | | Crystal structure of full-length FixJ from B. japonicum crystallized in space group C2221 | | Descriptor: | FORMIC ACID, GLYCEROL, Response regulator FixJ | | Authors: | Nishizono, Y, Hisano, T, Sawai, H, Shiro, Y, Nakamura, H, Wright, G.S.A, Saeki, A, Hikima, T, Yamamoto, M, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.778 Å) | | Cite: | Architecture of the complete oxygen-sensing FixL-FixJ two-component signal transduction system.
Sci Signal, 11, 2018
|
|
5XT2
 
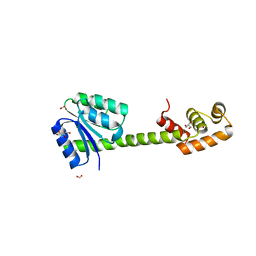 | | Crystal structures of full-length FixJ from B. japonicum crystallized in space group P212121 | | Descriptor: | FORMIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Nishizono, Y, Hisano, T, Shiro, Y, Sawai, H, Wright, G.S.A, Saeki, A, Hikima, T, Nakamura, H, Yamamoto, M, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-16 | | Release date: | 2018-05-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.652 Å) | | Cite: | Architecture of the complete oxygen-sensing FixL-FixJ two-component signal transduction system.
Sci Signal, 11, 2018
|
|
1ZRM
 
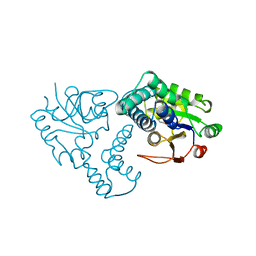 | | CRYSTAL STRUCTURE OF THE REACTION INTERMEDIATE OF L-2-HALOACID DEHALOGENASE WITH 2-CHLORO-N-BUTYRATE | | Descriptor: | L-2-HALOACID DEHALOGENASE, butanoic acid | | Authors: | Li, Y.-F, Hata, Y, Fujii, T, Hisano, T, Nishihara, M, Kurihara, T, Esaki, N. | | Deposit date: | 1998-03-03 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of reaction intermediates of L-2-haloacid dehalogenase and implications for the reaction mechanism.
J.Biol.Chem., 273, 1998
|
|
7W7C
 
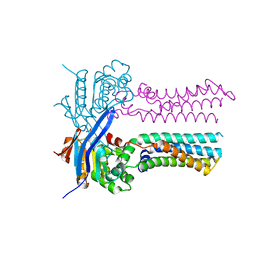 | | Heme exporter in the unliganded form | | Descriptor: | Putative ABC transport system integral membrane protein, Putative ABC transport system, ATP-binding protein, ... | | Authors: | Rahman, M.M, Hisano, T, Nakamura, H, Tosha, T, Shirouzu, M, Shiro, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2ECR
 
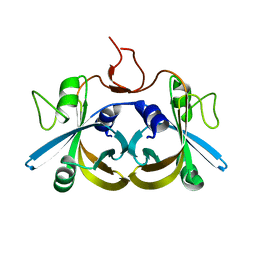 | | Crystal structure of the ligand-free form of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase | | Descriptor: | flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase | | Authors: | Kim, S.H, Hisano, T, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-02-13 | | Release date: | 2008-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8: Structural basis for the flavin affinity
Proteins, 70, 2008
|
|
