1DJ1
 
 | | CRYSTAL STRUCTURE OF R48A MUTANT OF CYTOCHROME C PEROXIDASE | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hirst, J, Goodin, D.B. | | Deposit date: | 1999-11-30 | | Release date: | 1999-12-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Unusual oxidative chemistry of N(omega)-hydroxyarginine and N-hydroxyguanidine catalyzed at an engineered cavity in a heme peroxidase.
J.Biol.Chem., 275, 2000
|
|
1DJ5
 
 | |
1DSO
 
 | | CYTOCHROME C PEROXIDASE H175G MUTANT, IMIDAZOLE COMPLEX AT PH 6, ROOM TEMPERATURE. | | Descriptor: | CYTOCHROME C PEROXIDASE, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hirst, J, Wilcox, S.K, Williams, P.A, McRee, D.E, Goodin, D.B. | | Deposit date: | 2000-01-07 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Replacement of the axial histidine ligand with imidazole in cytochrome c peroxidase. 1. Effects on structure.
Biochemistry, 40, 2001
|
|
1DSE
 
 | | CYTOCHROME C PEROXIDASE H175G MUTANT, IMIDAZOLE COMPLEX, WITH PHOSPHATE BOUND, PH 6, 100K | | Descriptor: | CYTOCHROME C PEROXIDASE, IMIDAZOLE, PHOSPHATE ION, ... | | Authors: | Hirst, J, Wilcox, S.K, Williams, P.A, McRee, D.E, Goodin, D.B. | | Deposit date: | 2000-01-07 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Replacement of the axial histidine ligand with imidazole in cytochrome c peroxidase. 1. Effects on structure.
Biochemistry, 40, 2001
|
|
1DS4
 
 | | CYTOCHROME C PEROXIDASE H175G MUTANT, IMIDAZOLE COMPLEX, PH 6, 100K | | Descriptor: | CYTOCHROME C PEROXIDASE, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hirst, J, Wilcox, S.K, Williams, P.A, McRee, D.E, Goodin, D.B. | | Deposit date: | 2000-01-07 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Replacement of the axial histidine ligand with imidazole in cytochrome c peroxidase. 1. Effects on structure.
Biochemistry, 40, 2001
|
|
1DSP
 
 | | CYTOCHROME C PEROXIDASE H175G MUTANT, IMIDAZOLE COMPLEX AT PH 7, ROOM TEMPERATURE. | | Descriptor: | CYTOCHROME C PEROXIDASE, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hirst, J, Wilcox, S.K, Williams, P.A, McRee, D.E, Goodin, D.B. | | Deposit date: | 2000-01-07 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Replacement of the axial histidine ligand with imidazole in cytochrome c peroxidase. 1. Effects on structure.
Biochemistry, 40, 2001
|
|
1DSG
 
 | | CYTOCHROME C PEROXIDASE H175G MUTANT, IMIDAZOLE COMPLEX AT PH 5, ROOM TEMPERATURE. | | Descriptor: | CYTOCHROME C PEROXIDASE, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hirst, J, Wilcox, S.K, Williams, P.A, McRee, D.E, Goodin, D.B. | | Deposit date: | 2000-01-07 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Replacement of the axial histidine ligand with imidazole in cytochrome c peroxidase. 1. Effects on structure.
Biochemistry, 40, 2001
|
|
6YJ4
 
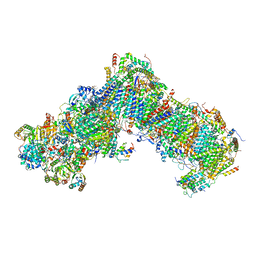 | | Structure of Yarrowia lipolytica complex I at 2.7 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Acyl carrier protein ACPM1 of NADH:Ubiquinone Oxidoreductase (Complex I), Acyl carrier protein ACPM2 of NADH:Ubiquinone Oxidoreductase (Complex I), ... | | Authors: | Hirst, J, Grba, D. | | Deposit date: | 2020-04-02 | | Release date: | 2020-08-12 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mitochondrial complex I structure reveals ordered water molecules for catalysis and proton translocation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7B0N
 
 | |
6YJ5
 
 | |
4UQ8
 
 | | Electron cryo-microscopy of bovine Complex I | | Descriptor: | ACYL CARRIER PROTEIN, MITOCHONDRIAL, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Vinothkumar, K.R, Zhu, J, Hirst, J. | | Deposit date: | 2014-06-21 | | Release date: | 2014-10-01 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4.95 Å) | | Cite: | Architecture of Mammalian Respiratory Complex I.
Nature, 515, 2014
|
|
1D3W
 
 | | Crystal structure of ferredoxin 1 d15e mutant from azotobacter vinelandii at 1.7 angstrom resolution. | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN 1, IRON/SULFUR CLUSTER | | Authors: | Chen, K, Hirst, J, Camba, R, Bonagura, C.A, Stout, C.D, Burges, B.K, Armstrong, F.A. | | Deposit date: | 1999-10-01 | | Release date: | 1999-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Atomically defined mechanism for proton transfer to a buried redox centre in a protein.
Nature, 405, 2000
|
|
2V07
 
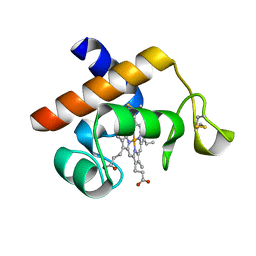 | | Structure of the Arabidopsis thaliana cytochrome c6A V52Q variant | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Worrall, J.A.R, Schlarb-Ridley, B.G, Reda, T, Marcaida, M.J, Moorlen, R.J, Wastl, J, Hirst, J, Bendall, D.S, Luisi, B.F, Howe, C.J. | | Deposit date: | 2007-05-09 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Modulation of heme redox potential in the cytochrome c6 family.
J. Am. Chem. Soc., 129, 2007
|
|
2V08
 
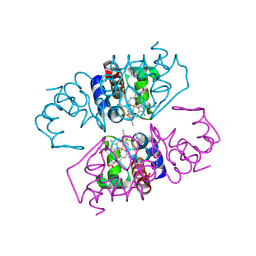 | | Structure of wild-type Phormidium laminosum cytochrome c6 | | Descriptor: | CHLORIDE ION, CYTOCHROME C6, HEME C, ... | | Authors: | Worrall, J.A.R, Schlarb-Ridley, B.G, Reda, T, Marcaida, M.J, Moorlen, R.J, Wastl, J, Hirst, J, Bendall, D.S, Luisi, B.F, Howe, C.J. | | Deposit date: | 2007-05-10 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Modulation of heme redox potential in the cytochrome c6 family.
J. Am. Chem. Soc., 129, 2007
|
|
5COD
 
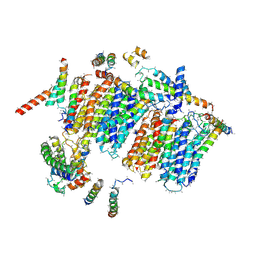 | | Bovine heart complex I membrane domain | | Descriptor: | NADH-ubiquinone oxidoreductase chain 4, NADH-ubiquinone oxidoreductase chain 5, SDAP, ... | | Authors: | Zhu, J, Hirst, J, King, M.S, Yu, M, Leslie, A.G.W, Klipcan, L. | | Deposit date: | 2015-07-20 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (6.74 Å) | | Cite: | Structure of subcomplex I beta of mammalian respiratory complex I leads to new supernumerary subunit assignments.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8OLT
 
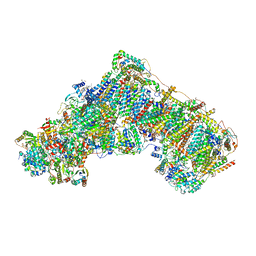 | | Mitochondrial complex I from Mus musculus in the active state bound with piericidin A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Grba, D.N, Chung, I, Bridges, H.R, Agip, A.N.A, Hirst, J. | | Deposit date: | 2023-03-30 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Investigation of hydrated channels and proton pathways in a high-resolution cryo-EM structure of mammalian complex I.
Sci Adv, 9, 2023
|
|
8OM1
 
 | | Mitochondrial complex I from Mus musculus in the active state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Grba, D.N, Chung, I, Bridges, H.R, Agip, A.N.A, Hirst, J. | | Deposit date: | 2023-03-31 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Investigation of hydrated channels and proton pathways in a high-resolution cryo-EM structure of mammalian complex I.
Sci Adv, 9, 2023
|
|
5LDX
 
 | | Structure of mammalian respiratory Complex I, class3. | | Descriptor: | Acyl carrier protein, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Vinothkumar, K.R, Zhu, J, Hirst, J. | | Deposit date: | 2016-06-28 | | Release date: | 2016-09-14 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structure of mammalian respiratory complex I.
Nature, 536, 2016
|
|
5LC5
 
 | | Structure of mammalian respiratory Complex I, class2 | | Descriptor: | Acyl carrier protein, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Vinothkumar, K.R, Zhu, J, Hirst, J. | | Deposit date: | 2016-06-19 | | Release date: | 2016-09-07 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Structure of mammalian respiratory complex I.
Nature, 536, 2016
|
|
5LDW
 
 | | Structure of mammalian respiratory Complex I, class1 | | Descriptor: | Acyl carrier protein, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Vinothkumar, K.R, Zhu, J, Hirst, J. | | Deposit date: | 2016-06-28 | | Release date: | 2016-09-07 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structure of mammalian respiratory complex I.
Nature, 536, 2016
|
|
8CA4
 
 | | Cryo-EM structure NDUFS4 knockout complex I from Mus musculus heart (Class 2 N-domain). | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Yin, Z, Bridges, H.R, Agip, A.N.A, Hirst, J. | | Deposit date: | 2023-01-24 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural insights into respiratory complex I deficiency and assembly from the mitochondrial disease-related ndufs4 -/- mouse.
Embo J., 43, 2024
|
|
8C2S
 
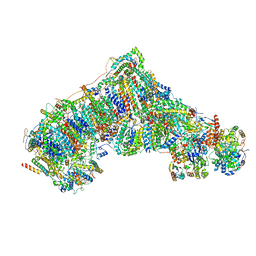 | | Cryo-EM structure NDUFS4 knockout complex I from Mus musculus heart (Class 1). | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Yin, Z, Bridges, H.R, Agip, A.N.A, Hirst, J. | | Deposit date: | 2022-12-22 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into respiratory complex I deficiency and assembly from the mitochondrial disease-related ndufs4 -/- mouse.
Embo J., 43, 2024
|
|
8CA3
 
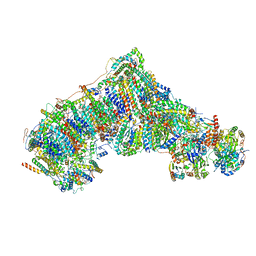 | | Cryo-EM structure NDUFS4 knockout complex I from Mus musculus heart (Class 2). | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Yin, Z, Bridges, H.R, Agip, A.N.A, Hirst, J. | | Deposit date: | 2023-01-24 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into respiratory complex I deficiency and assembly from the mitochondrial disease-related ndufs4 -/- mouse.
Embo J., 43, 2024
|
|
8CA1
 
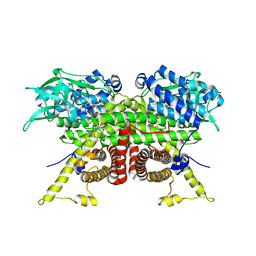 | | Cryo-EM structure of the ACADVL dimer from Mus musculus. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Very long-chain specific acyl-CoA dehydrogenase, mitochondrial | | Authors: | Yin, Z, Bridges, H.R, Agip, A.N.A, Hirst, J. | | Deposit date: | 2023-01-24 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural insights into respiratory complex I deficiency and assembly from the mitochondrial disease-related ndufs4 -/- mouse.
Embo J., 43, 2024
|
|
8CA5
 
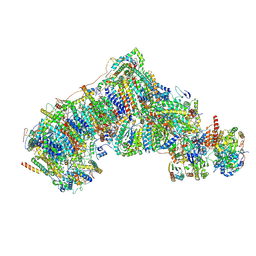 | | Cryo-EM structure NDUFS4 knockout complex I from Mus musculus heart (Class 3). | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Yin, Z, Bridges, H.R, Agip, A.N.A, Hirst, J. | | Deposit date: | 2023-01-24 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into respiratory complex I deficiency and assembly from the mitochondrial disease-related ndufs4 -/- mouse.
Embo J., 43, 2024
|
|
