6W1X
 
 | | Cryo-EM structure of anti-CRISPR AcrIF9, bound to the type I-F crRNA-guided CRISPR surveillance complex | | Descriptor: | CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, CRISPR-associated protein Csy3, ... | | Authors: | Hirschi, M, Santiago-Frangos, A, Wilkinson, R, Golden, S.M, Wiedenheft, B, Lander, G. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | AcrIF9 tethers non-sequence specific dsDNA to the CRISPR RNA-guided surveillance complex.
Nat Commun, 11, 2020
|
|
6WHI
 
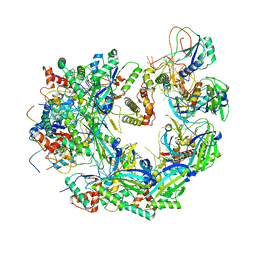 | | Cryo-electron microscopy structure of the type I-F CRISPR RNA-guided surveillance complex bound to the anti-CRISPR AcrIF9 | | Descriptor: | CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, CRISPR-associated protein Csy3, ... | | Authors: | Hirschi, M, Santiago-Frangos, A, Wilkinson, R, Golden, S.M, Wiedenheft, B, Lander, G. | | Deposit date: | 2020-04-08 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | AcrIF9 tethers non-sequence specific dsDNA to the CRISPR RNA-guided surveillance complex.
Nat Commun, 11, 2020
|
|
5U9W
 
 | | Structure of CNTnw N149L in the intermediate 3 state | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-beta-D-glucopyranosyl)oxy]methyl}-2-octyldecyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, Nucleoside permease | | Authors: | Hirschi, M, Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2016-12-18 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.555 Å) | | Cite: | Visualizing multistep elevator-like transitions of a nucleoside transporter.
Nature, 545, 2017
|
|
5W3S
 
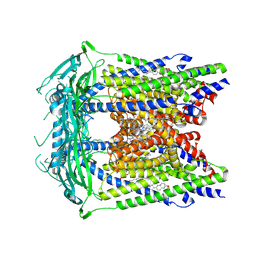 | | Cryo-electron microscopy structure of a TRPML3 ion channel | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL HEMISUCCINATE, Mucolipin-3 isoform 1, ... | | Authors: | Hirschi, M, Herzik, M.A, Wie, J, Suo, Y, Borschel, W.F, Ren, D, Lander, G.C, Lee, S.Y. | | Deposit date: | 2017-06-08 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Cryo-electron microscopy structure of the lysosomal calcium-permeable channel TRPML3.
Nature, 550, 2017
|
|
5L26
 
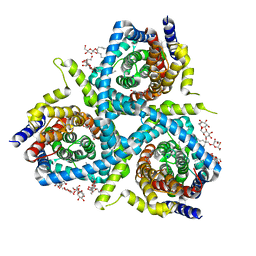 | | Structure of CNTnw in an inward-facing substrate-bound state | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-beta-D-glucopyranosyl)oxy]methyl}-2-octyldecyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, Nucleoside permease, SODIUM ION, ... | | Authors: | Hirschi, M, Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2016-07-31 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Visualizing multistep elevator-like transitions of a nucleoside transporter.
Nature, 545, 2017
|
|
5L24
 
 | | Structure of CNTnw N149L in the intermediate 2 state | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-beta-D-glucopyranosyl)oxy]methyl}-2-octyldecyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, Nucleoside permease | | Authors: | Hirschi, M, Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2016-07-31 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Visualizing multistep elevator-like transitions of a nucleoside transporter.
Nature, 545, 2017
|
|
5L27
 
 | | Structure of CNTnw N149L in the intermediate 1 state | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-beta-D-glucopyranosyl)oxy]methyl}-2-octyldecyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, Nucleoside permease | | Authors: | Hirschi, M, Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2016-07-31 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Visualizing multistep elevator-like transitions of a nucleoside transporter.
Nature, 545, 2017
|
|
5L2B
 
 | | Structure of CNTnw N149S, E332A in an outward-facing state | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-beta-D-glucopyranosyl)oxy]methyl}-2-octyldecyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, Nucleoside permease | | Authors: | Hirschi, M, Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2016-07-31 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Visualizing multistep elevator-like transitions of a nucleoside transporter.
Nature, 545, 2017
|
|
5L2A
 
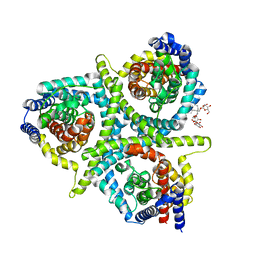 | | Structure of CNTnw N149S,F366A in an outward-facing state | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-beta-D-glucopyranosyl)oxy]methyl}-2-octyldecyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, Nucleoside permease | | Authors: | Hirschi, M, Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2016-07-31 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Visualizing multistep elevator-like transitions of a nucleoside transporter.
Nature, 545, 2017
|
|
8FFY
 
 | | Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA(UGA-TL) | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, Serine--tRNA ligase, mitochondrial, ... | | Authors: | Hirschi, M, Kuhle, B, Doerfel, L, Schimmel, P, Lander, G. | | Deposit date: | 2022-12-11 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for a degenerate tRNA identity code and the evolution of bimodal specificity in human mitochondrial tRNA recognition.
Nat Commun, 14, 2023
|
|
8VQ4
 
 | | CDK2-CyclinE1 in complex with allosteric inhibitor I-125A. | | Descriptor: | (8R)-6-(1-benzyl-1H-pyrazole-4-carbonyl)-N-[(2S,3R)-3-(2-cyclohexylethoxy)-1-(methylamino)-1-oxobutan-2-yl]-2-[(1S)-2,2-dimethylcyclopropane-1-carbonyl]-2,6-diazaspiro[3.4]octane-8-carboxamide, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Hirschi, M, Johnson, E, Zhang, Y, Liu, Z, Brodsky, O, Won, S.J, Nagata, A, Petroski, M.D, Majmudar, J.D, Niessen, S, VanArsdale, T, Gilbert, A.M, Hayward, M.M, Stewart, A.E, Nager, A.R, Melillo, B, Cravatt, B. | | Deposit date: | 2024-01-17 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Expanding the ligandable proteome by paralog hopping with covalent probes.
Biorxiv, 2024
|
|
8VQ3
 
 | | CDK2-CyclinE1 in complex with allosteric inhibitor I-198. | | Descriptor: | (8R)-N-[(2S,3R)-3-(cyclohexylmethoxy)-1-(morpholin-4-yl)-1-oxobutan-2-yl]-2-[(1S)-2,2-dimethylcyclopropane-1-carbonyl]-6-(1,3-thiazole-5-carbonyl)-2,6-diazaspiro[3.4]octane-8-carboxamide, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Hirschi, M, Johnson, E, Zhang, Y, Liu, Z, Brodsky, O, Won, S.J, Nagata, A, Petroski, M.D, Majmudar, J.D, Niessen, S, VanArsdale, T, Gilbert, A.M, Hayward, M.M, Stewart, A.E, Nager, A.R, Melillo, B, Cravatt, B. | | Deposit date: | 2024-01-17 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Expanding the ligandable proteome by paralog hopping with covalent probes.
Biorxiv, 2024
|
|
7U2A
 
 | | Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA (GCU) | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, RNA (38-MER), Serine--tRNA ligase, ... | | Authors: | Hirschi, M, Kuhle, B, Doerfel, L, Schimmel, P, Lander, G. | | Deposit date: | 2022-02-23 | | Release date: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for shape-selective recognition and aminoacylation of a D-armless human mitochondrial tRNA.
Nat Commun, 13, 2022
|
|
7U2B
 
 | | Cryo-electron microscopy structure of human mt-SerRS in complex with mt-tRNA(GCU-TL) | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, RNA (53-MER), Serine--tRNA ligase, ... | | Authors: | Hirschi, M, Kuhle, B, Doerfel, L, Schimmel, P, Lander, G. | | Deposit date: | 2022-02-23 | | Release date: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for shape-selective recognition and aminoacylation of a D-armless human mitochondrial tRNA.
Nat Commun, 13, 2022
|
|
7TZB
 
 | | Crystal structure of the human mitochondrial seryl-tRNA synthetase (mt SerRS) bound with a seryl-adenylate analogue | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, Serine--tRNA ligase, mitochondrial | | Authors: | Kuhle, B, Hirschi, M, Doerfel, L, Lander, G, Schimmel, P. | | Deposit date: | 2022-02-15 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for shape-selective recognition and aminoacylation of a D-armless human mitochondrial tRNA.
Nat Commun, 13, 2022
|
|
6MHS
 
 | | Structure of the human TRPV3 channel in a putative sensitized conformation | | Descriptor: | Transient receptor potential cation channel subfamily V member 3 | | Authors: | Zubcevic, L, Herzik, M.A, Wu, M, Borschel, W.F, Hirschi, M, Song, A, Lander, G.C, Lee, S.Y. | | Deposit date: | 2018-09-18 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational ensemble of the human TRPV3 ion channel.
Nat Commun, 9, 2018
|
|
6MHX
 
 | | Structure of human TRPV3 in the presence of 2-APB in C2 symmetry (2) | | Descriptor: | Transient receptor potential cation channel subfamily V member 3 | | Authors: | Zubcevic, L, Herzik, M.A, Wu, M, Borschel, W.F, Hirschi, M, Song, A, Lander, G.C, Lee, S.Y. | | Deposit date: | 2018-09-18 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational ensemble of the human TRPV3 ion channel.
Nat Commun, 9, 2018
|
|
6MHW
 
 | | Structure of human TRPV3 in the presence of 2-APB in C2 symmetry (1) | | Descriptor: | Transient receptor potential cation channel subfamily V member 3 | | Authors: | Zubcevic, L, Herzik, M.A, Wu, M, Borschel, W.F, Hirschi, M, Song, A, Lander, G.C, Lee, S.Y. | | Deposit date: | 2018-09-18 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational ensemble of the human TRPV3 ion channel.
Nat Commun, 9, 2018
|
|
6MHV
 
 | | Structure of human TRPV3 in the presence of 2-APB in C4 symmetry | | Descriptor: | Transient receptor potential cation channel subfamily V member 3 | | Authors: | Zubcevic, L, Herzik, M.A, Wu, M, Borschel, W.F, Hirschi, M, Song, A, Lander, G.C, Lee, S.Y. | | Deposit date: | 2018-09-18 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformational ensemble of the human TRPV3 ion channel.
Nat Commun, 9, 2018
|
|
6MHO
 
 | | Structure of the human TRPV3 channel in the apo conformation | | Descriptor: | Transient receptor potential cation channel subfamily V member 3 | | Authors: | Zubcevic, L, Herzik, M.A, Wu, M, Borschel, W.F, Hirschi, M, Song, A, Lander, G.C, Lee, S.Y. | | Deposit date: | 2018-09-18 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Conformational ensemble of the human TRPV3 ion channel.
Nat Commun, 9, 2018
|
|
3RHK
 
 | | Crystal structure of the catalytic domain of c-Met kinase in complex with ARQ 197 | | Descriptor: | 1-[(3R,4R)-4-(1H-indol-3-yl)-2,5-dioxopyrrolidin-3-yl]pyrrolo[3,2,1-ij]quinolinium, Hepatocyte growth factor receptor | | Authors: | Eathiraj, S, Palma, R, Volckova, E, Hirschi, M, France, D.S, Ashwell, M.A, Chan, T.C. | | Deposit date: | 2011-04-11 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of a novel mode of protein kinase inhibition characterized by the mechanism of inhibition of human mesenchymal-epithelial transition factor (c-Met) protein autophosphorylation by ARQ 197.
J.Biol.Chem., 286, 2011
|
|
3RHX
 
 | | Crystal structure of the catalytic domain of FGFR1 kinase in complex with ARQ 069 | | Descriptor: | (6S)-6-phenyl-5,6-dihydrobenzo[h]quinazolin-2-amine, 1,2-ETHANEDIOL, Basic fibroblast growth factor receptor 1, ... | | Authors: | Eathiraj, S, Palma, R, Hirschi, M, Volckova, E, Nakuci, E, Castro, J, Chen, C.R, Chan, T.C, France, D.S, Ashwell, M.A. | | Deposit date: | 2011-04-12 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A novel mode of protein kinase inhibition exploiting hydrophobic motifs of autoinhibited kinases: discovery of ATP-independent inhibitors of fibroblast growth factor receptor.
J.Biol.Chem., 286, 2011
|
|
3RI1
 
 | | Crystal structure of the catalytic domain of FGFR2 kinase in complex with ARQ 069 | | Descriptor: | (6S)-6-phenyl-5,6-dihydrobenzo[h]quinazolin-2-amine, Fibroblast growth factor receptor 2, SULFATE ION | | Authors: | Eathiraj, S, Palma, R, Hirschi, M, Volckova, E, Nakuci, E, Castro, J, Chen, C.R, Chan, T.C, France, D.S, Ashwell, M.A. | | Deposit date: | 2011-04-12 | | Release date: | 2011-05-04 | | Last modified: | 2011-10-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A novel mode of protein kinase inhibition exploiting hydrophobic motifs of autoinhibited kinases: discovery of ATP-independent inhibitors of fibroblast growth factor receptor.
J.Biol.Chem., 286, 2011
|
|
