1V4G
 
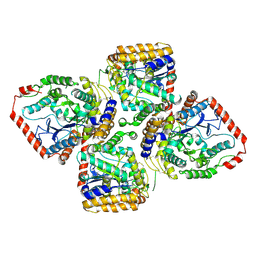 | | Crystal Structure of gamma-Glutamylcysteine Synthetase from Escherichia coli B | | Descriptor: | Glutamate--cysteine ligase | | Authors: | Hibi, T, Nii, H, Nakatsu, T, Kato, H, Hiratake, J, Oda, J. | | Deposit date: | 2003-11-13 | | Release date: | 2004-10-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of gamma-glutamylcysteine synthetase: insights into the mechanism of catalysis by a key enzyme for glutathione homeostasis
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
1VA6
 
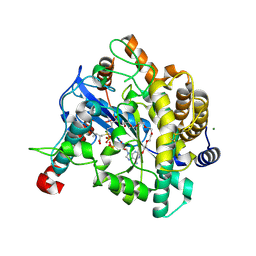 | | Crystal structure of Gamma-glutamylcysteine synthetase from Escherichia Coli B complexed with Transition-state analogue | | Descriptor: | (2S)-2-AMINO-4-[[(2R)-2-CARBOXYBUTYL](PHOSPHONO)SULFONIMIDOYL]BUTANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, Glutamate--cysteine ligase, ... | | Authors: | Hibi, T, Nii, H, Nakatsu, T, Kato, H, Hiratake, J, Oda, J. | | Deposit date: | 2004-02-12 | | Release date: | 2004-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of gamma-glutamylcysteine synthetase: insights into the mechanism of catalysis by a key enzyme for glutathione homeostasis
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
2D1Q
 
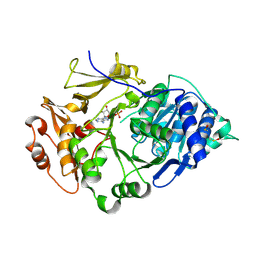 | | Crystal structure of the thermostable Japanese Firefly Luciferase complexed with MgATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Luciferin 4-monooxygenase | | Authors: | Nakatsu, T, Ichiyama, S, Hiratake, J, Saldanha, A, Kobashi, N, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-31 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the spectral difference in luciferase bioluminescence.
Nature, 440, 2006
|
|
2D1R
 
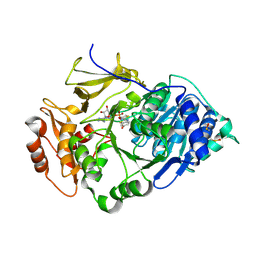 | | Crystal structure of the thermostable Japanese firefly Luciferase complexed with OXYLUCIFERIN and AMP | | Descriptor: | 2-(6-HYDROXY-1,3-BENZOTHIAZOL-2-YL)-1,3-THIAZOL-4(5H)-ONE, ADENOSINE MONOPHOSPHATE, Luciferin 4-monooxygenase | | Authors: | Nakatsu, T, Ichiyama, S, Hiratake, J, Saldanha, A, Kobashi, N, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-31 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the spectral difference in luciferase bioluminescence.
Nature, 440, 2006
|
|
2D1S
 
 | | Crystal structure of the thermostable Japanese Firefly Luciferase complexed with High-energy intermediate analogue | | Descriptor: | 5'-O-[N-(DEHYDROLUCIFERYL)-SULFAMOYL] ADENOSINE, CHLORIDE ION, Luciferin 4-monooxygenase | | Authors: | Nakatsu, T, Ichiyama, S, Hiratake, J, Saldanha, A, Kobashi, N, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-31 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for the spectral difference in luciferase bioluminescence.
Nature, 440, 2006
|
|
2D1T
 
 | | Crystal structure of the thermostable Japanese Firefly Luciferase red-color emission S286N mutant complexed with High-energy intermediate analogue | | Descriptor: | 5'-O-[N-(DEHYDROLUCIFERYL)-SULFAMOYL] ADENOSINE, CHLORIDE ION, Luciferin 4-monooxygenase | | Authors: | Nakatsu, T, Ichiyama, S, Hiratake, J, Saldanha, A, Kobashi, N, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-31 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for the spectral difference in luciferase bioluminescence.
Nature, 440, 2006
|
|
3WHR
 
 | | Crystal structure of gamma-glutamyltranspeptidase from Bacillus subtilis (crystal soaked for 3min. in acivicin soln. ) | | Descriptor: | Gamma-glutamyltranspeptidase large chain, Gamma-glutamyltranspeptidase small chain | | Authors: | Ida, T, Suzuki, H, Fukuyama, K, Hiratake, J, Wada, K. | | Deposit date: | 2013-08-30 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of Bacillus subtilis gamma-glutamyltranspeptidase in complex with acivicin: diversity of the binding mode of a classical and electrophilic active-site-directed glutamate analogue.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5B5T
 
 | | Crystal Structure of Escherichia coli Gamma-Glutamyltranspeptidase in Complex with peptidyl phosphonate inhibitor 1b | | Descriptor: | (2~{S})-2-azanyl-4-[(2~{R})-1-(2-hydroxy-2-oxoethylamino)-1-oxidanylidene-butan-2-yl]oxyphosphonoyl-butanoic acid, CALCIUM ION, Gamma-glutamyltranspeptidase large chain, ... | | Authors: | Wada, K, Fukuyama, K. | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Phosphonate-based irreversible inhibitors of human gamma-glutamyl transpeptidase (GGT). GGsTop is a non-toxic and highly selective inhibitor with critical electrostatic interaction with an active-site residue Lys562 for enhanced inhibitory activity
Bioorg.Med.Chem., 24, 2016
|
|
2GLT
 
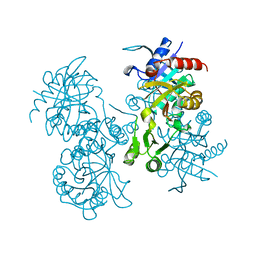 | | STRUCTURE OF ESCHERICHIA COLI GLUTATHIONE SYNTHETASE AT PH 6.0. | | Descriptor: | GLUTATHIONE BIOSYNTHETIC LIGASE | | Authors: | Matsuda, K, Yamaguchi, H, Kato, H, Nishioka, T, Katsube, Y, Oda, J. | | Deposit date: | 1995-05-16 | | Release date: | 1995-07-31 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glutathione synthetase at optimal pH: domain architecture and structural similarity with other proteins.
Protein Eng., 9, 1996
|
|
1GSH
 
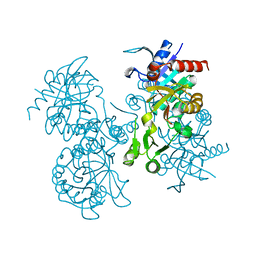 | | STRUCTURE OF ESCHERICHIA COLI GLUTATHIONE SYNTHETASE AT PH 7.5 | | Descriptor: | GLUTATHIONE BIOSYNTHETIC LIGASE | | Authors: | Matsuda, K, Kato, H, Yamaguchi, H, Nishioka, T, Katsube, Y, Oda, J. | | Deposit date: | 1995-05-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of glutathione synthetase at optimal pH: domain architecture and structural similarity with other proteins.
Protein Eng., 9, 1996
|
|
2Z8I
 
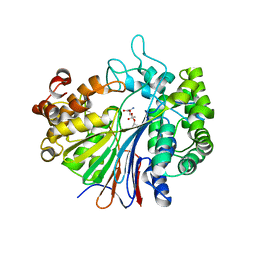 | |
2Z8K
 
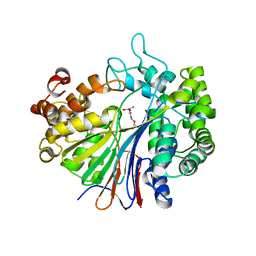 | |
2Z8J
 
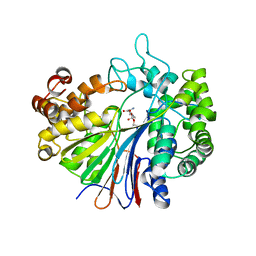 | |
3WMG
 
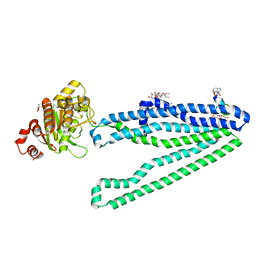 | | Crystal structure of an inward-facing eukaryotic ABC multidrug transporter G277V/A278V/A279V mutant in complex with an cyclic peptide inhibitor, aCAP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ATP-binding cassette, sub-family B, ... | | Authors: | Kodan, A, Yamaguchi, T, Nakatsu, T, Sakiyama, K, Hipolito, C.J, Fujioka, A, Hirokane, R, Ikeguchi, K, Watanabe, B, Hirtake, J, Kimura, Y, Suga, H, Ueda, K, Kato, H. | | Deposit date: | 2013-11-18 | | Release date: | 2014-04-30 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for gating mechanisms of a eukaryotic P-glycoprotein homolog.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2D32
 
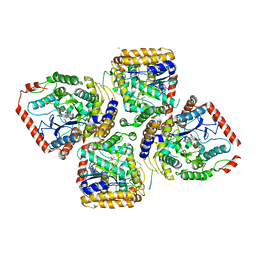 | | Crystal Structure of Michaelis Complex of gamma-Glutamylcysteine Synthetase | | Descriptor: | CYSTEINE, GLUTAMIC ACID, Glutamate--cysteine ligase, ... | | Authors: | Hibi, T, Nakayama, M, Nii, H, Kurokawa, Y, Katano, H, Oda, J. | | Deposit date: | 2005-09-25 | | Release date: | 2006-11-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of efficient coupling peptide ligation and ATP hydrolysis by gamma-gluatamylcysteine synthetase
To be Published
|
|
2D33
 
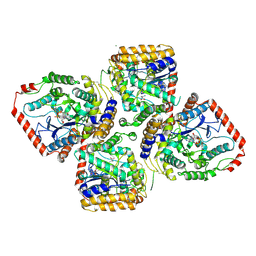 | | Crystal Structure of gamma-Glutamylcysteine Synthetase Complexed with Aluminum Fluoride | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CYSTEINE, ... | | Authors: | Hibi, T, Nakayama, M, Nii, H, Kurokawa, Y, Katano, H, Oda, J. | | Deposit date: | 2005-09-25 | | Release date: | 2006-11-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of efficient coupling between peptide ligation and ATP hydrolysis by gamma-gluatamylcysteine synthetase
To be Published
|
|
3WQ4
 
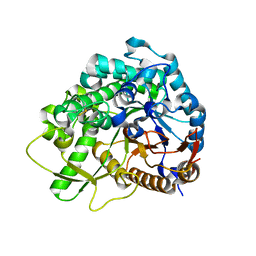 | | Crystal structure of beta-primeverosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-primeverosidase | | Authors: | Saino, H. | | Deposit date: | 2014-01-22 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of beta-primeverosidase in complex with disaccharide amidine inhibitors.
J.Biol.Chem., 289, 2014
|
|
3WHQ
 
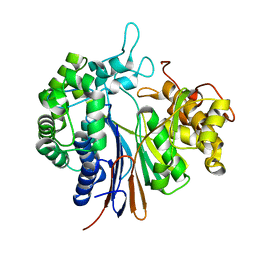 | |
3WHS
 
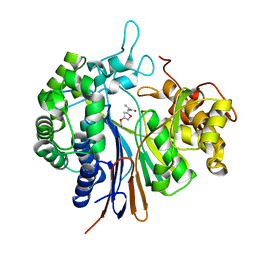 | | Crystal structure of Bacillus subtilis gamma-glutamyltranspeptidase in complex with acivicin | | Descriptor: | (2S)-AMINO[(5S)-3-CHLORO-4,5-DIHYDROISOXAZOL-5-YL]ACETIC ACID, Gamma-glutamyltranspeptidase large chain, Gamma-glutamyltranspeptidase small chain | | Authors: | Wada, K, Fukuyama, K. | | Deposit date: | 2013-08-30 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Bacillus subtilis gamma-glutamyltranspeptidase in complex with acivicin: diversity of the binding mode of a classical and electrophilic active-site-directed glutamate analogue.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WQ5
 
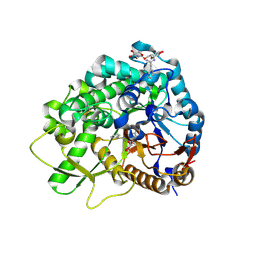 | |
3WME
 
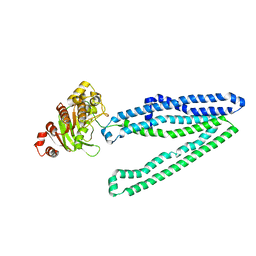 | | Crystal structure of an inward-facing eukaryotic ABC multidrug transporter | | Descriptor: | ATP-binding cassette, sub-family B, member 1, ... | | Authors: | Kodan, A, Yamaguchi, T, Nakatsu, T, Kato, H. | | Deposit date: | 2013-11-18 | | Release date: | 2014-03-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Structural basis for gating mechanisms of a eukaryotic P-glycoprotein homolog.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3WQ6
 
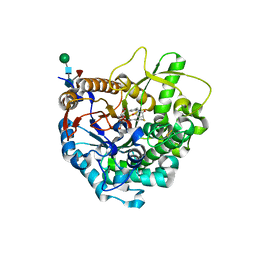 | |
3WMF
 
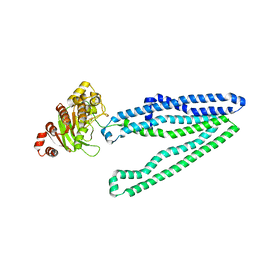 | | Crystal structure of an inward-facing eukaryotic ABC multitrug transporter G277V/A278V/A279V mutant | | Descriptor: | ATP-binding cassette, sub-family B, member 1, ... | | Authors: | Kodan, A, Yamaguchi, T, Nakatsu, T, Kato, H. | | Deposit date: | 2013-11-18 | | Release date: | 2014-03-19 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for gating mechanisms of a eukaryotic P-glycoprotein homolog.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2ZXQ
 
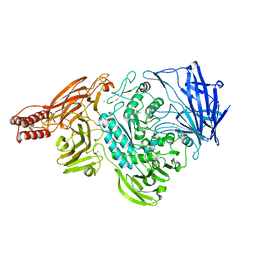 | | Crystal structure of endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum (EngBF) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Endo-alpha-N-acetylgalactosaminidase, MANGANESE (II) ION | | Authors: | Suzuki, R, Katayama, T, Ashida, H, Yamamoto, K, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2009-01-05 | | Release date: | 2009-06-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and mutational analyses of substrate recognition of endo-{alpha}-N-acetylgalactosaminidase from Bifidobacterium longum.
J.Biochem., 2009
|
|
