5FLE
 
 | | High resolution NI,FE-CODH-320 mV with CN state | | Descriptor: | CARBON MONOXIDE DEHYDROGENASE 2, FE (II) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Ciaccafava, A, Tombolelli, D, Domnik, L, Fesseler, J, Jeoung, J.-H, Dobbek, H, Mroginski, M.A, Hildebrandt, P, Zebger, I. | | Deposit date: | 2015-10-26 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | When the inhibitor tells more than the substrate: the cyanide-bound state of a carbon monoxide dehydrogenase.
Chem Sci, 7, 2016
|
|
7ZXV
 
 | | Orange Carotenoid Protein Trp-288 BTA mutant | | Descriptor: | CHLORIDE ION, Orange carotenoid-binding protein, beta,beta-caroten-4-one, ... | | Authors: | Moldenhauer, M, Tseng, H.-W, Kraskov, A, Tavraz, N.N, Hildebrandt, P, Hochberg, G, Essen, L.-O, Budisa, N, Korf, L, Maksimov, E.G, Friedrich, T. | | Deposit date: | 2022-05-23 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Parameterization of a single H-bond in Orange Carotenoid Protein by atomic mutation reveals principles of evolutionary design of complex chemical photosystems.
Front Mol Biosci, 10, 2023
|
|
2WSD
 
 | | Proximal mutations at the type 1 Cu site of CotA-laccase: I494A mutant | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, OXYGEN MOLECULE, ... | | Authors: | Silva, C.S, Durao, P, Chen, Z, Soares, C.M, Pereira, M.M, Todorovic, S, Hildebrandt, P, Martins, L.O, Lindley, P.F, Bento, I. | | Deposit date: | 2009-09-04 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Proximal Mutations at the Type 1 Copper Site of Cota Laccase: Spectroscopic, Redox, Kinetic and Structural Characterization of I494A and L386A Mutants.
Biochem.J., 412, 2008
|
|
4AKQ
 
 | | Mutations in the neighbourhood of CotA-laccase trinuclear site: E498D mutant | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, OXYGEN MOLECULE, ... | | Authors: | Silva, C.S, Chen, Z, Durao, P, Pereira, M.M, Todorovic, S, Hildebrandt, P, Martins, L.O, Lindley, P.F, Bento, I. | | Deposit date: | 2012-02-28 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Role of Glu498 in the Dioxygen Reactivity of Cota-Laccase from Bacillus Subtilis.
Dalton Trans, 39, 2010
|
|
4AKP
 
 | | Mutations in the neighbourhood of CotA-laccase trinuclear site: E498T mutant | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, OXYGEN MOLECULE, ... | | Authors: | Silva, C.S, Chen, Z, Durao, P, Pereira, M.M, Todorovic, S, Hildebrandt, P, Martins, L.O, Lindley, P.F, Bento, I. | | Deposit date: | 2012-02-28 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Role of Glu498 in the Dioxygen Reactivity of Cota-Laccase from Bacillus Subtilis.
Dalton Trans, 39, 2010
|
|
4AKO
 
 | | Mutations in the neighbourhood of CotA-laccase trinuclear site: E498L mutant | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, OXYGEN MOLECULE, ... | | Authors: | Silva, C.S, Chen, Z, Durao, P, Pereira, M.M, Todorovic, S, Hildebrandt, P, Martins, L.O, Lindley, P.F, Bento, I. | | Deposit date: | 2012-02-28 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Role of Glu498 in the Dioxygen Reactivity of Cota-Laccase from Bacillus Subtilis.
Dalton Trans, 39, 2010
|
|
4C1N
 
 | | Corrinoid protein reactivation complex with activator | | Descriptor: | CARBON MONOXIDE DEHYDROGENASE CORRINOID/IRON-SULFUR PROTEIN, GAMMA SUBUNIT, CO DEHYDROGENASE/ACETYL-COA SYNTHASE, ... | | Authors: | Hennig, S.E, Goetzl, S, Jeoung, J.H, Bommer, M, Lendzian, F, Hildebrandt, P, Dobbek, H. | | Deposit date: | 2013-08-13 | | Release date: | 2014-08-13 | | Last modified: | 2014-08-20 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | ATP-Induced Electron Transfer by Redox-Selective Partner Recognition
Nat.Commun., 5, 2014
|
|
7ODH
 
 | |
7ODG
 
 | |
4UTI
 
 | | XenA - oxidized - Y183F variant in complex with coumarin | | Descriptor: | COUMARIN, FLAVIN MONONUCLEOTIDE, NADH:flavin oxidoreductase, ... | | Authors: | Werther, T, Dobbek, H. | | Deposit date: | 2014-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.099 Å) | | Cite: | Redox-dependent substrate-cofactor interactions in the Michaelis-complex of a flavin-dependent oxidoreductase
Nat Commun, 8, 2017
|
|
4UTL
 
 | | XenA - reduced - Y183F variant in complex with coumarin | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, COUMARIN, SULFATE ION, ... | | Authors: | Werther, T, Dobbek, H. | | Deposit date: | 2014-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.229 Å) | | Cite: | Redox-dependent substrate-cofactor interactions in the Michaelis-complex of a flavin-dependent oxidoreductase
Nat Commun, 8, 2017
|
|
4UTJ
 
 | |
4UTK
 
 | | XenA - reduced - Y183F variant | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, SULFATE ION, XENOBIOTIC REDUCTASE | | Authors: | Werther, T, Dobbek, H. | | Deposit date: | 2014-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Redox-dependent substrate-cofactor interactions in the Michaelis-complex of a flavin-dependent oxidoreductase
Nat Commun, 8, 2017
|
|
4IUD
 
 | | Crystal structure of an O2-tolerant [NiFe]-hydrogenase from Ralstonia eutropha in its as-isolated form with ascorbate - partly reduced state | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Hammer, M, Schmidt, A, Frielingsdorf, S, Fritsch, J, Lenz, O, Scheerer, P. | | Deposit date: | 2013-01-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Reversible [4Fe-3S] cluster morphing in an O2-tolerant [NiFe] hydrogenase.
Nat.Chem.Biol., 10, 2014
|
|
4IUC
 
 | | Crystal structure of an O2-tolerant [NiFe]-hydrogenase from Ralstonia eutropha in its as-isolated form - oxidized state 2 | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Frielingsdorf, S, Schmidt, A, Fritsch, J, Lenz, O, Scheerer, P. | | Deposit date: | 2013-01-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Reversible [4Fe-3S] cluster morphing in an O2-tolerant [NiFe] hydrogenase.
Nat.Chem.Biol., 10, 2014
|
|
4IUB
 
 | | Crystal structure of an O2-tolerant [NiFe]-hydrogenase from Ralstonia eutropha in its as-isolated form - oxidized state 1 | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Frielingsdorf, S, Schmidt, A, Fritsch, J, Lenz, O, Scheerer, P. | | Deposit date: | 2013-01-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Reversible [4Fe-3S] cluster morphing in an O2-tolerant [NiFe] hydrogenase.
Nat.Chem.Biol., 10, 2014
|
|
4UTM
 
 | | XenA - Reduced - Y183F variant in complex with 8-hydroxycoumarin | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 8-HYDROXYCOUMARIN, SULFATE ION, ... | | Authors: | Werther, T, Dobbek, H. | | Deposit date: | 2014-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Redox-dependent substrate-cofactor interactions in the Michaelis-complex of a flavin-dependent oxidoreductase
Nat Commun, 8, 2017
|
|
4UTH
 
 | | XenA - oxidized - Y183F variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADH:flavin oxidoreductase, SULFATE ION | | Authors: | Werther, T, Dobbek, H. | | Deposit date: | 2014-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Redox-dependent substrate-cofactor interactions in the Michaelis-complex of a flavin-dependent oxidoreductase
Nat Commun, 8, 2017
|
|
5LNI
 
 | |
5LNJ
 
 | | XenA - reduced - Y183F variant in complex with 7-hydroxycoumarin | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 7-hydroxy-2H-chromen-2-one, NADH:flavin oxidoreductase, ... | | Authors: | Werther, T, Dobbek, H. | | Deposit date: | 2016-08-04 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Redox-dependent substrate-cofactor interactions in the Michaelis-complex of a flavin-dependent oxidoreductase
Nat Commun, 8, 2017
|
|
5AKP
 
 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP from Xanthomonas campestris bound to BV chromophore | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BILIVERDINE IX ALPHA, CHLORIDE ION, ... | | Authors: | Otero, L.H, Klinke, S, Goldbaum, F.A, Bonomi, H.R. | | Deposit date: | 2015-03-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the Full-Length Bacteriophytochrome from the Plant Pathogen Xanthomonas Campestris Provides Clues to its Long-Range Signaling Mechanism.
J.Mol.Biol., 428, 2016
|
|
6G1Y
 
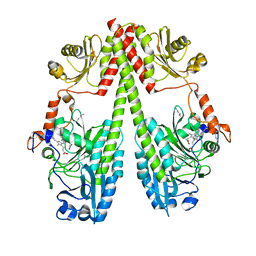 | | Crystal structure of the photosensory core module (PCM) of a bathy phytochrome from Agrobacterium fabrum in the Pfr state. | | Descriptor: | 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, Bacteriophytochrome protein | | Authors: | Schmidt, A, Qureshi, B.M, Scheerer, P. | | Deposit date: | 2018-03-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural snapshot of a bacterial phytochrome in its functional intermediate state.
Nat Commun, 9, 2018
|
|
6G20
 
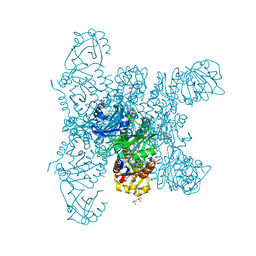 | | Crystal structure of a fluorescence optimized bathy phytochrome PAiRFP2 derived from wild-type Agp2 in its functional Meta-F intermediate state. | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Schmidt, A, Sauthof, L, Szczepek, M, Scheerer, P. | | Deposit date: | 2018-03-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural snapshot of a bacterial phytochrome in its functional intermediate state.
Nat Commun, 9, 2018
|
|
6G1Z
 
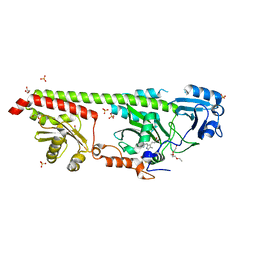 | | Crystal structure of a fluorescence optimized bathy phytochrome PAiRFP2 derived from wild-type Agp2 in its Pfr state. | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Sauthof, L, Schmidt, A, Szczepek, M, Scheerer, P. | | Deposit date: | 2018-03-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural snapshot of a bacterial phytochrome in its functional intermediate state.
Nat Commun, 9, 2018
|
|
6SAW
 
 | |
