1BQF
 
 | | GROWTH-BLOCKING PEPTIDE (GBP) FROM PSEUDALETIA SEPARATA | | Descriptor: | PROTEIN (GROWTH-BLOCKING PEPTIDE) | | Authors: | Aizawa, T, Fujitani, N, Hayakawa, Y, Ohnishi, A, Ohkubo, T, Kwano, K, Hikichi, K, Nitta, K. | | Deposit date: | 1998-08-09 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an insect growth factor, growth-blocking peptide.
J.Biol.Chem., 274, 1999
|
|
1LKJ
 
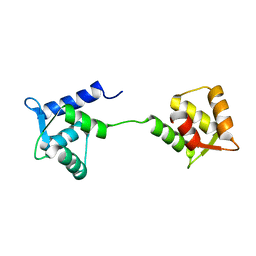 | | NMR Structure of Apo Calmodulin from Yeast Saccharomyces cerevisiae | | Descriptor: | Calmodulin | | Authors: | Ishida, H, Nakashima, K, Kumaki, Y, Nakata, M, Hikichi, K, Yazawa, M. | | Deposit date: | 2002-04-25 | | Release date: | 2003-04-29 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of apocalmodulin from Saccharomyces cerevisiae implies a mechanism for its unique Ca2+ binding property.
Biochemistry, 41, 2002
|
|
1F54
 
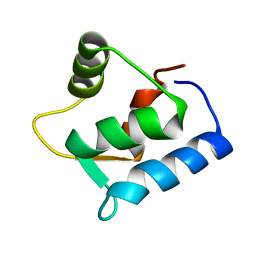 | | SOLUTION STRUCTURE OF THE APO N-TERMINAL DOMAIN OF YEAST CALMODULIN | | Descriptor: | CALMODULIN | | Authors: | Ishida, H, Takahashi, K, Nakashima, K, Kumaki, Y, Nakata, M, Hikichi, K, Yazawa, M. | | Deposit date: | 2000-06-13 | | Release date: | 2003-07-15 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the N-terminal Domain of Yeast Calmodulin:
Ca2+-Dependent Conformational Change and Its Functional Implication
Biochemistry, 39, 2000
|
|
1F55
 
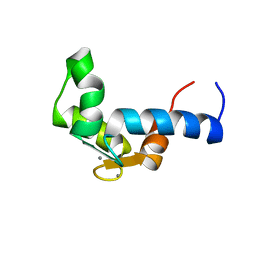 | | SOLUTION STRUCTURE OF THE CALCIUM BOUND N-TERMINAL DOMAIN OF YEAST CALMODULIN | | Descriptor: | CALCIUM ION, CALMODULIN | | Authors: | Ishida, H, Takahashi, K, Nakashima, K, Kumaki, Y, Nakata, M, Hikichi, K, Yazawa, M. | | Deposit date: | 2000-06-13 | | Release date: | 2003-07-15 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the N-terminal Domain of Yeast Calmodulin:
Ca2+-Dependent Conformational Change and Its Functional Implication
Biochemistry, 39, 2000
|
|
1DQC
 
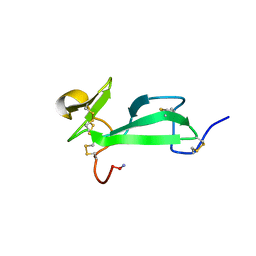 | | SOLUTION STRUCTURE OF TACHYCITIN, AN ANTIMICROBIAL PROTEIN WITH CHITIN-BINDING FUNCTION | | Descriptor: | TACHYCITIN | | Authors: | Suetake, T, Tsuda, S, Kawabata, S, Miura, K, Kawano, K. | | Deposit date: | 2000-01-04 | | Release date: | 2000-09-13 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Chitin-binding proteins in invertebrates and plants comprise a common chitin-binding structural motif.
J.Biol.Chem., 275, 2000
|
|
3NLA
 
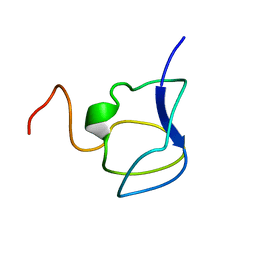 | | NMR STRUCTURE OF THE N-TERMINAL DOMAIN WITH A LINKER PORTION OF ANTARCTIC EEL POUT ANTIFREEZE PROTEIN RD3, 40 STRUCTURES | | Descriptor: | ANTIFREEZE PROTEIN RD3 TYPE III | | Authors: | Miura, K, Ohgiya, S, Hoshino, T, Nemoto, N, Hikichi, K, Tsuda, S. | | Deposit date: | 1998-02-24 | | Release date: | 1999-02-23 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of a globular antifreeze protein to ice.
Nature, 384, 1996
|
|
3RDN
 
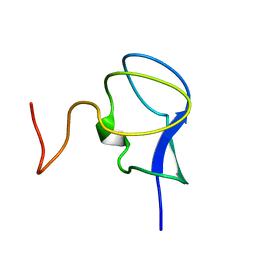 | | NMR STRUCTURE OF THE N-TERMINAL DOMAIN WITH A LINKER PORTION OF ANTARCTIC EEL POUT ANTIFREEZE PROTEIN RD3, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | ANTIFREEZE PROTEIN RD3 TYPE III | | Authors: | Miura, K, Ohgiya, S, Hoshino, T, Nemoto, N, Hikichi, K, Tsuda, S. | | Deposit date: | 1998-02-24 | | Release date: | 1999-02-23 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of a globular antifreeze protein to ice.
Nature, 384, 1996
|
|
