4AXV
 
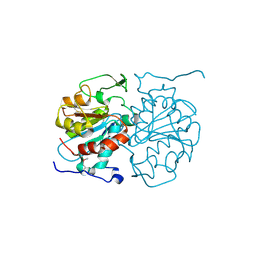 | | Biochemical and structural characterization of the MpaA amidase as part of a conserved scavenging pathway for peptidoglycan derived peptides in gamma-proteobacteria | | Descriptor: | 1,2-ETHANEDIOL, MPAA, ZINC ION | | Authors: | Maqbool, A, Herve, M, Mengin-Lecreulx, D, Dodson, E, Wilkinson, A.J, Thomas, G.H. | | Deposit date: | 2012-06-14 | | Release date: | 2012-09-26 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Mpaa is a Murein-Tripeptide-Specific Zinc Carboxypeptidase that Functions as Part of a Catabolic Pathway for Peptidoglycan Derived Peptides in Gamma-Proteobacteria.
Biochem.J., 448, 2012
|
|
2JFH
 
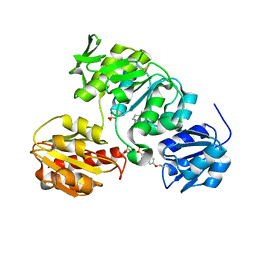 | | Crystal structure of MurD ligase in complex with L-Glu containing sulfonamide inhibitor | | Descriptor: | N-[(6-BUTOXYNAPHTHALEN-2-YL)SULFONYL]-L-GLUTAMIC ACID, SULFATE ION, UDP-N-ACETYLMURAMOYL-ALANINE-D-GLUTAMATE LIGASE | | Authors: | Kotnik, M, Humljan, J, Contreras-Martel, C, Oblak, M, Kristan, K, Herve, M, Blanot, D, Urleb, U, Gobec, S, Dessen, A, Solmajer, T. | | Deposit date: | 2007-02-01 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Functional Characterization of Enantiomeric Glutamic Acid Derivatives as Potential Transition State Analogue Inhibitors of Murd Ligase.
J.Mol.Biol., 370, 2007
|
|
2JFF
 
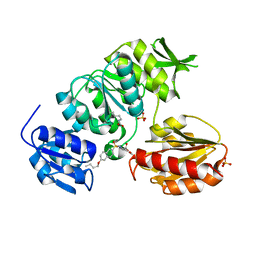 | | Crystal structure of MurD ligase in complex with D-Glu containing sulfonamide inhibitor | | Descriptor: | N-[(6-BUTOXYNAPHTHALEN-2-YL)SULFONYL]-D-GLUTAMIC ACID, SULFATE ION, UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE | | Authors: | Kotnik, M, Humljan, J, Contreras-Martel, C, Oblak, M, Kristan, K, Herve, M, Blanot, D, Urleb, U, Gobec, S, Dessen, A, Solmajer, T. | | Deposit date: | 2007-02-01 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and Functional Characterization of Enantiomeric Glutamic Acid Derivatives as Potential Transition State Analogue Inhibitors of Murd Ligase.
J.Mol.Biol., 370, 2007
|
|
2JFG
 
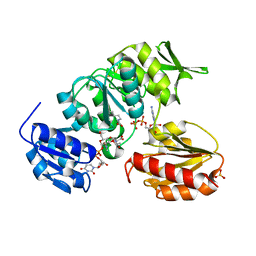 | | Crystal structure of MurD ligase in complex with UMA and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE, ... | | Authors: | Kotnik, M, Humljan, J, Contreras-Martel, C, Oblak, M, Kristan, K, Herve, M, Blanot, D, Urleb, U, Gobec, S, Dessen, A, Solmajer, T. | | Deposit date: | 2007-02-01 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and functional characterization of enantiomeric glutamic acid derivatives as potential transition state analogue inhibitors of MurD ligase.
J. Mol. Biol., 370, 2007
|
|
3G23
 
 | |
1OE7
 
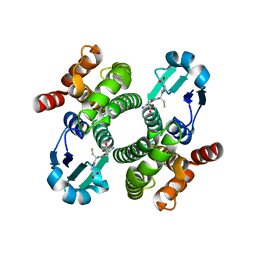 | |
1OE8
 
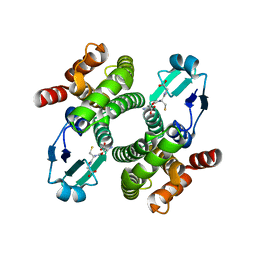 | |
3HN7
 
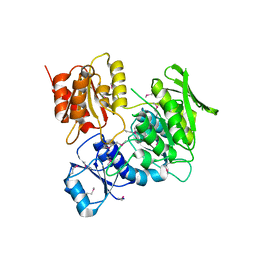 | |
4QDN
 
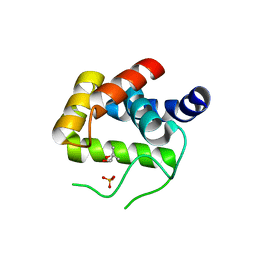 | | Crystal Structure of the endo-beta-N-acetylglucosaminidase from Thermotoga maritima | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Flagellar protein FlgJ [peptidoglycan hydrolase], PHOSPHATE ION | | Authors: | Lipski, A, Nurizzo, D, Bourne, Y, Vincent, F. | | Deposit date: | 2014-05-14 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical characterization of the beta-N-acetylglucosaminidase from Thermotoga maritima: Toward rationalization of mechanistic knowledge in the GH73 family.
Glycobiology, 25, 2015
|
|
4R9S
 
 | | Mycobacterium tuberculosis InhA bound to NITD-916 | | Descriptor: | 6-[(4,4-dimethylcyclohexyl)methyl]-4-hydroxy-3-phenylpyridin-2(1H)-one, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Noble, C.G. | | Deposit date: | 2014-09-07 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Direct inhibitors of InhA are active against Mycobacterium tuberculosis
Sci Transl Med, 7, 2015
|
|
4R9R
 
 | | Mycobacterium tuberculosis InhA bound to NITD-564 | | Descriptor: | 6-(cyclohexylmethyl)-4-hydroxy-3-phenylpyridin-2(1H)-one, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Noble, C.G. | | Deposit date: | 2014-09-07 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Direct inhibitors of InhA are active against Mycobacterium tuberculosis
Sci Transl Med, 7, 2015
|
|
3M9G
 
 | | Crystal structure of the three-PASTA-domain of a Ser/Thr kinase from Staphylococcus aureus | | Descriptor: | Protein kinase, ZINC ION | | Authors: | Paracuellos, P, Ballandras, A, Robert, X, Creze, C, Cozzone, A.J, Duclos, B, Gouet, P. | | Deposit date: | 2010-03-22 | | Release date: | 2010-11-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Extended Conformation of the 2.9-A Crystal Structure of the Three-PASTA Domain of a Ser/Thr Kinase from the Human Pathogen Staphylococcus aureus
J.Mol.Biol., 404, 2010
|
|
3O9P
 
 | | The structure of the Escherichia coli murein tripeptide binding protein MppA | | Descriptor: | L-ALA-GAMMA-D-GLU-MESO-DIAMINOPIMELIC ACID, Periplasmic murein peptide-binding protein, ZINC ION | | Authors: | Maqbool, A, Levdikov, V.M, Blagova, E.V, Wilkinson, A.J, Thomas, G.H. | | Deposit date: | 2010-08-04 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Compensating Stereochemical Changes Allow Murein Tripeptide to Be Accommodated in a Conventional Peptide-binding Protein.
J.Biol.Chem., 286, 2011
|
|
2F8F
 
 | | Crystal structure of the Y10F mutant of the gluathione s-transferase from schistosoma haematobium | | Descriptor: | GLUTATHIONE, Glutathione S-transferase 28 kDa | | Authors: | Gourlay, L.J, Baiocco, P, Angelucci, F, Miele, A.E, Bellelli, A, Brunori, M. | | Deposit date: | 2005-12-02 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the Mechanism of GSH Activation in Schistosoma haematobium Glutathione-S-transferase by Site-directed Mutagenesis and X-ray Crystallography.
J.Mol.Biol., 360, 2006
|
|
2C80
 
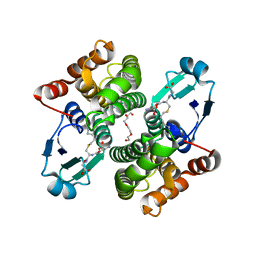 | | Structure of Sh28GST in complex with S-hexyl Glutathione | | Descriptor: | GLUTATHIONE S-TRANSFERASE 28 KDA, S-HEXYLGLUTATHIONE, TETRAETHYLENE GLYCOL | | Authors: | Baiocco, P, Gourlay, L.J, Angelucci, F, Bellelli, A, Brunori, M, Miele, A.E. | | Deposit date: | 2005-11-30 | | Release date: | 2006-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the Mechanism of Gsh Activation in Schistosoma Haematobium Glutathione-S-Transferase by Site-Directed Mutagenesis and X-Ray Crystallography.
J.Mol.Biol., 360, 2006
|
|
2CAI
 
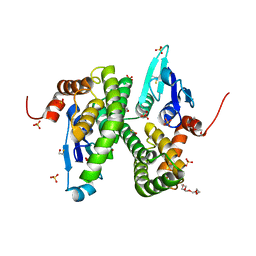 | | Structure of Glutathione-S-Transferase mutant, R21L, from Schistosoma Haematobium | | Descriptor: | BETA-MERCAPTOETHANOL, GLUTATHIONE S-TRANSFERASE 28 KDA, SULFATE ION, ... | | Authors: | Baiocco, P, Gourlay, L.J, Angelucci, F, Bellelli, A, Brunori, M, Miele, A.E. | | Deposit date: | 2005-12-21 | | Release date: | 2006-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Probing the Mechanism of Gsh Activation in Schistosoma Haematobium Glutathione-S-Transferase by Site-Directed Mutagenesis and X-Ray Crystallography.
J.Mol.Biol., 360, 2006
|
|
2C8U
 
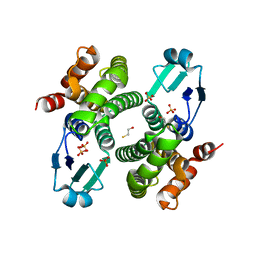 | | Structure of R21Q mutant of Sh28GST | | Descriptor: | BETA-MERCAPTOETHANOL, GLUTATHIONE S-TRANSFERASE 28 KDA, SULFATE ION | | Authors: | Baiocco, P, Gourlay, L.J, Angelucci, F, Bellelli, A, Miele, A.E, Brunori, M. | | Deposit date: | 2005-12-07 | | Release date: | 2006-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the Mechanism of Gsh Activation in Schistosoma Haematobium Glutathione-S-Transferase by Site-Directed Mutagenesis and X-Ray Crystallography.
J.Mol.Biol., 360, 2006
|
|
2CA8
 
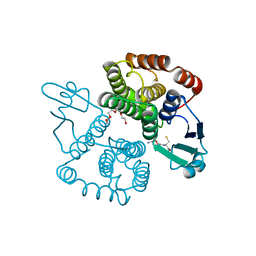 | | Structure of Sh28GST in complex with GSH at pH 6.0 | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE 28 KDA, TETRAETHYLENE GLYCOL | | Authors: | Baiocco, P, Gourlay, L.J, Angelucci, F, Bellelli, A, Miele, A.E, Brunori, M. | | Deposit date: | 2005-12-20 | | Release date: | 2006-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Probing the Mechanism of Gsh Activation in Schistosoma Haematobium Glutathione-S-Transferase by Site-Directed Mutagenesis and X-Ray Crystallography.
J.Mol.Biol., 360, 2006
|
|
2CAQ
 
 | | Structure of R21L mutant of Sh28GST in complex with GSH | | Descriptor: | BETA-MERCAPTOETHANOL, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE 28 KDA, ... | | Authors: | Baiocco, P, Gourlay, L.J, Angelucci, F, Bellelli, A, Miele, A.E, Brunori, M. | | Deposit date: | 2005-12-22 | | Release date: | 2006-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the Mechanism of Gsh Activation in Schistosoma Haematobium Glutathione-S-Transferase by Site-Directed Mutagenesis and X-Ray Crystallography.
J.Mol.Biol., 360, 2006
|
|
1S2J
 
 | | Crystal structure of the Drosophila pattern-recognition receptor PGRP-SA | | Descriptor: | PHOSPHATE ION, Peptidoglycan recognition protein SA CG11709-PA | | Authors: | Chang, C.-I, Pili-Floury, S, Chelliah, Y, Lemaitre, B, Mengin-Lecreulx, D, Deisenhofer, J. | | Deposit date: | 2004-01-08 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Drosophila pattern recognition receptor contains a peptidoglycan docking groove and unusual l,d-carboxypeptidase activity.
PLOS BIOL., 2, 2004
|
|
