7VEN
 
 | | Structure of burosumab Fab | | Descriptor: | Burosumab Fab HC, Burosumab Fab LC | | Authors: | Heo, Y.S. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of the Fab Fragment of Burosumab
To Be Published
|
|
8GSI
 
 | | Structure of the cobolimab Fab | | Descriptor: | heavy chain, light chain, nanobody | | Authors: | Heo, Y.S, Choi, S.B. | | Deposit date: | 2022-09-06 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structure of the Fab fragment of cobolimab, an investigational antibody targeting Tim-3 for immune-oncology.
To Be Published
|
|
4HQ6
 
 | | BC domain in the presence of citrate | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Heo, Y.S. | | Deposit date: | 2012-10-25 | | Release date: | 2013-10-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights into the Regulation of ACC2 by Citrate
Bull.Korean Chem.Soc., 34, 2013
|
|
5GGU
 
 | | Crystal structure of tremelimumab Fab | | Descriptor: | heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2016-11-16 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5GGS
 
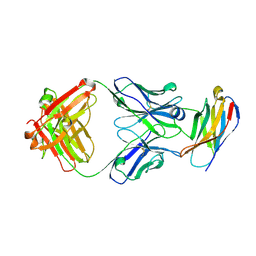 | | PD-1 in complex with pembrolizumab Fab | | Descriptor: | Programmed cell death protein 1, heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2016-11-16 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5GGV
 
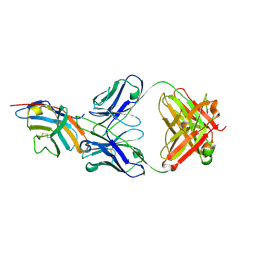 | | CTLA-4 in complex with tremelimumab Fab | | Descriptor: | Cytotoxic T-lymphocyte protein 4, heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2016-11-16 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5GGT
 
 | | PD-L1 in complex with BMS-936559 Fab | | Descriptor: | IGK@ protein, IgG H chain, Programmed cell death 1 ligand 1 | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5GGR
 
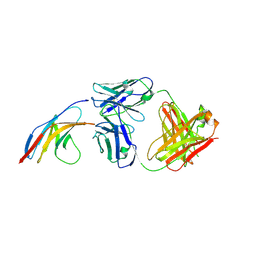 | | PD-1 in complex with nivolumab Fab | | Descriptor: | Programmed cell death protein 1, heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2016-11-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5GGQ
 
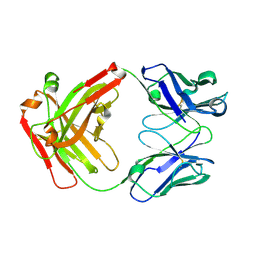 | | Crystal structure of Nivolumab Fab fragment | | Descriptor: | nivolumab heavy chain, nivolumab light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2016-11-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
3LPX
 
 | | Crystal structure of GyrA | | Descriptor: | DNA gyrase, A subunit | | Authors: | Jung, H.Y, Heo, Y.S. | | Deposit date: | 2010-02-07 | | Release date: | 2011-02-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of GyrA
To be Published
|
|
7WSL
 
 | | PD-1 in complex with Dostarlimab | | Descriptor: | Programmed cell death protein 1, heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2022-01-30 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Molecular basis of PD-1 blockade by dostarlimab, the FDA-approved antibody for cancer immunotherapy.
Biochem.Biophys.Res.Commun., 599, 2022
|
|
7BXA
 
 | | Crystal structure of PD-1 in complex with tislelizumab Fab | | Descriptor: | Programmed cell death protein 1, heavy chain, light chain | | Authors: | Heo, Y.S, Lee, S.H, Lim, H, Lee, H.T, Kim, Y.J, Park, E.B. | | Deposit date: | 2020-04-18 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Crystal structure of PD-1 in complex with an antibody-drug tislelizumab used in tumor immune checkpoint therapy.
Biochem.Biophys.Res.Commun., 527, 2020
|
|
6JC2
 
 | | Crystal structure of the Fab fragment of ipilimumab | | Descriptor: | SULFATE ION, ipilimumab fab heavy chain, ipilimumab fab light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2019-01-27 | | Release date: | 2019-09-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of the Fab Fragment of an Anti-CTLA-4 Antibody, Ipilimumab, Used for Cancer Immunotherapy
Bull.Korean Chem.Soc., 40, 2019
|
|
5WUX
 
 | | TNFalpha-certolizumab Fab | | Descriptor: | Tumor necrosis factor alpha, heavy, light | | Authors: | Heo, Y.S, Lee, J.U. | | Deposit date: | 2016-12-21 | | Release date: | 2017-06-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Basis for the Neutralization of Tumor Necrosis Factor alpha by Certolizumab Pegol in the Treatment of Inflammatory Autoimmune Diseases
Int J Mol Sci, 18, 2017
|
|
5WUV
 
 | | Crystal structure of Certolizumab Fab | | Descriptor: | heavy chain, light chain | | Authors: | Heo, Y.S, Lee, J.U, Son, J.Y, Shin, W, Yoo, K.Y. | | Deposit date: | 2016-12-21 | | Release date: | 2017-06-07 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Molecular Basis for the Neutralization of Tumor Necrosis Factor alpha by Certolizumab Pegol in the Treatment of Inflammatory Autoimmune Diseases
Int J Mol Sci, 18, 2017
|
|
5X8M
 
 | | PD-L1 in complex with durvalumab | | Descriptor: | Programmed cell death 1 ligand 1, durvalumab heavy chain, durvalumab light chain | | Authors: | Heo, Y.S, Lee, H.T. | | Deposit date: | 2017-03-03 | | Release date: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (2.661 Å) | | Cite: | Molecular mechanism of PD-1/PD-L1 blockade via anti-PD-L1 antibodies atezolizumab and durvalumab
Sci Rep, 7, 2017
|
|
5X8L
 
 | | PD-L1 in complex with atezolizumab | | Descriptor: | Programmed cell death 1 ligand 1, atezolizumab heavy chain, atezolizumab light chain | | Authors: | Heo, Y.S, Lee, H.T. | | Deposit date: | 2017-03-03 | | Release date: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular mechanism of PD-1/PD-L1 blockade via anti-PD-L1 antibodies atezolizumab and durvalumab
Sci Rep, 7, 2017
|
|
2HJW
 
 | | Crystal Structure of the BC domain of ACC2 | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Cho, Y.S, Lee, J.I, Shin, D, Kim, H.T, Lee, T.G, Heo, Y.S. | | Deposit date: | 2006-07-02 | | Release date: | 2007-07-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the biotin carboxylase domain of human acetyl-CoA carboxylase 2.
Proteins, 70, 2008
|
|
2O5K
 
 | | Crystal Structure of GSK3beta in complex with a benzoimidazol inhibitor | | Descriptor: | 2-(2,4-DICHLORO-PHENYL)-7-HYDROXY-1H-BENZOIMIDAZOLE-4-CARBOXYLIC ACID [2-(4-METHANESULFONYLAMINO-PHENYL)-ETHYL]-AMIDE, Glycogen synthase kinase-3 beta | | Authors: | Shin, D, Lee, S.C, Heo, Y.S, Cho, Y.S, Kim, Y.E, Hyun, Y.L, Cho, J.M, Lee, Y.S, Ro, S. | | Deposit date: | 2006-12-06 | | Release date: | 2007-10-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design and synthesis of 7-hydroxy-1H-benzoimidazole derivatives as novel inhibitors of glycogen synthase kinase-3beta
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4Y7I
 
 | | Crystal Structure of MTMR8 | | Descriptor: | Myotubularin-related protein 8, PHOSPHATE ION | | Authors: | Yoo, K, Lee, J, Son, J, Shin, W, Im, D, Heo, Y.S. | | Deposit date: | 2015-02-15 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure of the catalytic phosphatase domain of MTMR8: implications for dimerization, membrane association and reversible oxidation.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3V3V
 
 | | Structural and functional analysis of quercetagetin, a natural JNK1 inhibitor | | Descriptor: | 3,5,6,7-TETRAHYDROXY-2-(3,4-DIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, C-Jun-amino-terminal kinase-interacting protein 1, CHLORIDE ION, ... | | Authors: | Baek, S, Kang, N.J, Popowicz, G.M, Arciniega, M, Jung, S.K, Byun, S, Song, N.R, Heo, Y.S, Kim, B.Y, Lee, H.J, Holak, T.A, Augustin, M, Bode, A.M, Huber, R, Dong, Z, Lee, K.W. | | Deposit date: | 2011-12-14 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Analysis of the Natural JNK1 Inhibitor Quercetagetin.
J.Mol.Biol., 425, 2013
|
|
3JRW
 
 | | Phosphorylated BC domain of ACC2 | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Cho, Y.S, Lee, J.I, Shin, D, Kim, H.T, Lee, T.G, Heo, Y.S. | | Deposit date: | 2009-09-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism for the regulation of human ACC2 through phosphorylation by AMPK
Biochem.Biophys.Res.Commun., 391, 2010
|
|
3JRX
 
 | | Crystal structure of the BC domain of ACC2 in complex with soraphen A | | Descriptor: | Acetyl-CoA carboxylase 2, SORAPHEN A | | Authors: | Cho, Y.S, Lee, J.I, Shin, D, Kim, H.T, Lee, T.G, Heo, Y.S. | | Deposit date: | 2009-09-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism for the regulation of human ACC2 through phosphorylation by AMPK.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
3NJ4
 
 | | Fluoro-neplanocin A in Human S-Adenosylhomocysteine Hydrolase | | Descriptor: | (4S,5S)-4-(6-amino-9H-purin-9-yl)-3-fluoro-5-hydroxy-2-(hydroxymethyl)cyclopent-2-en-1-one, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jeong, L.S, Lee, K.M, Hwang, K.Y, Choi, S, Heo, Y.S. | | Deposit date: | 2010-06-17 | | Release date: | 2011-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure and binding mode analysis of human S-adenosylhomocysteine hydrolase complexed with novel mechanism-based inhibitors, haloneplanocin A analogues.
J.Med.Chem., 54, 2011
|
|
1L8O
 
 | | Molecular basis for the local conformational rearrangement of human phosphoserine phosphatase | | Descriptor: | L-3-phosphoserine phosphatase, PHOSPHATE ION, SERINE | | Authors: | Kim, H.Y, Heo, Y.S, Kim, J.H, Park, M.H, Moon, J, Park, S.Y, Lee, T.G, Jeon, Y.H, Ro, S, Hwang, K.Y. | | Deposit date: | 2002-03-21 | | Release date: | 2003-04-01 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for the local conformational rearrangement of human phosphoserine phosphatase
J.Biol.Chem., 277, 2002
|
|
