4UTS
 
 | | Room temperature crystal structure of the fast switching M159T mutant of fluorescent protein Dronpa | | Descriptor: | FLUORESCENT PROTEIN DRONPA | | Authors: | Kaucikas, M, Fitzpatrick, A, Bryan, E, Struve, A, Henning, R, Kosheleva, I, Srajer, V, van Thor, J.J. | | Deposit date: | 2014-07-22 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Room Temperature Crystal Structure of the Fast Switching M159T Mutant of the Fluorescent Protein Dronpa.
Proteins, 83, 2015
|
|
6MH6
 
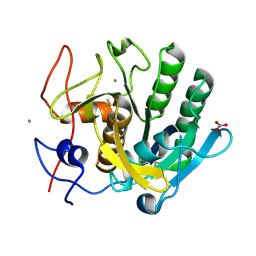 | | High-viscosity injector-based Pink Beam Serial Crystallography of Micro-crystals at a Synchrotron Radiation Source. | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Martin-Garcia, J.M, Zhu, L, Mendez, D, Lee, M, Chun, E, Li, C, Hu, H, Subramanian, G, Kissick, D, Ogata, C, Henning, R, Ishchenko, A, Dobson, Z, Zhan, S, Weierstall, U, Spence, J.C.H, Fromme, P, Zatsepin, N.A, Fischetti, R.F, Cherezov, V, Liu, W. | | Deposit date: | 2018-09-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-viscosity injector-based pink-beam serial crystallography of microcrystals at a synchrotron radiation source.
Iucrj, 6, 2019
|
|
6MH8
 
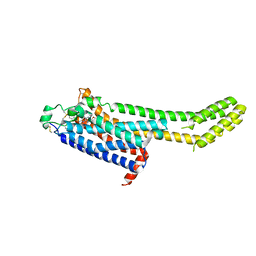 | | High-viscosity injector-based Pink Beam Serial Crystallography of Micro-crystals at a Synchrotron Radiation Source | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a, Soluble cytochrome b562 chimeric construct | | Authors: | Martin-Garcia, J.M, Zhu, L, Mendez, D, Lee, M, Chun, E, Li, C, Hu, H, Subramanian, G, Kissick, D, Ogata, C, Henning, R, Ishchenko, A, Dobson, Z, Zhan, S, Weierstall, U, Spence, J.C.H, Fromme, P, Zatsepin, N.A, Fischetti, R.F, Cherezov, V, Liu, W. | | Deposit date: | 2018-09-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | High-viscosity injector-based pink-beam serial crystallography of microcrystals at a synchrotron radiation source.
Iucrj, 6, 2019
|
|
7KOA
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/16 Methyltransferase in a Complex with Cap-0 and SAM Determined by Pink-Beam Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Shuvalova, L, Lavens, A, Henning, R, Maltseva, N, Rosas-Lemus, M, Kim, Y, Satchell, K.J.F, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with Cap-0 and SAM Determined by Pink-Beam Serial Crystallography
To Be Published
|
|
8CTV
 
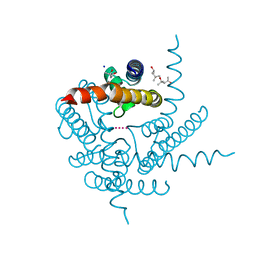 | | Crystal structure of a K+ selective NaK mutant (NaK2K) -Tl+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Potassium channel protein, SODIUM ION, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CTN
 
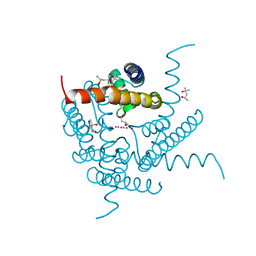 | | Structure of a K+ selective NaK mutant (NaK2K, Laue diffraction, no electric field) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CTW
 
 | | Crystal structure of a K+ selective NaK mutant (NaK2K) -Na+,Tl+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Potassium channel protein, SODIUM ION, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CTS
 
 | | Room temperature crystal structure of a K+ selective NaK mutant (NaK2K) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CTT
 
 | | Crystal structure of a K+ selective NaK mutant (NaK2K) at 100K | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CTX
 
 | | Crystal structure of a K+ selective NaK mutant (NaK2K) -K+,Tl+ complex | | Descriptor: | POTASSIUM ION, Potassium channel protein, THALLIUM (I) ION | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CTU
 
 | | Crystal structure of a K+ selective NaK mutant (NaK2K) at Room temperature | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CU1
 
 | | Structure of a K+ selective NaK mutant (NaK2K, Laue diffraction) in the presence of an electric field of ~0.8MV/cm along the crystallographic z axis, 500ns, with eightfold extrapolation of structure factor differences | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CU2
 
 | | Structure of a K+ selective NaK mutant (NaK2K, Laue diffraction) in the presence of an electric field of ~0.8MV/cm along the crystallographic z axis, 100ns, with eightfold extrapolation of structure factor differences | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CU4
 
 | | Structure of a K+ selective NaK mutant (NaK2K, Laue diffraction) in the presence of an electric field of ~0.8MV/cm along the crystallographic z axis, 1us, with eightfold extrapolation of structure factor differences | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
8CU3
 
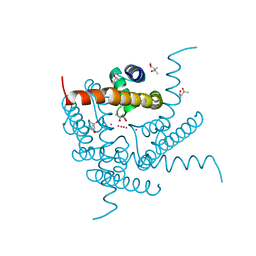 | | Structure of a K+ selective NaK mutant (NaK2K, Laue diffraction) in the presence of an electric field of ~0.8MV/cm along the crystallographic z axis, 200ns, with eightfold extrapolation of structure factor differences | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Lee, B, White, K.I, Socolich, M.A, Klureza, M.A, Henning, R, Srajer, V, Ranganathan, R, Hekstra, D. | | Deposit date: | 2022-05-16 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Direct visualization of electric field-stimulated ion conduction in a potassium channel
To Be Published
|
|
3UMD
 
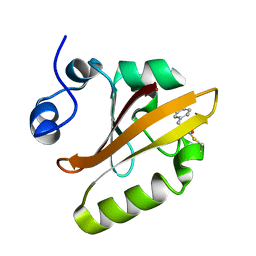 | | Structure of pB intermediate of Photoactive yellow protein (PYP) at pH 4. | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Tripathi, S, Srajer, V, Purwar, N, Henning, R, Schmidt, M. | | Deposit date: | 2011-11-13 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | pH Dependence of the Photoactive Yellow Protein Photocycle Investigated by Time-Resolved Crystallography.
Biophys.J., 102, 2012
|
|
3UME
 
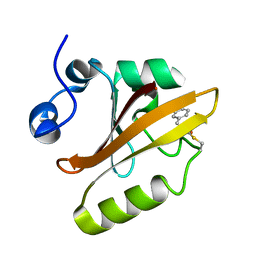 | | Structure of pB intermediate of Photoactive yellow protein (PYP) at pH 7 | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Tripathi, S, Srajer, V, Purwar, N, Henning, R, Schmidt, M. | | Deposit date: | 2011-11-13 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | pH Dependence of the Photoactive Yellow Protein Photocycle Investigated by Time-Resolved Crystallography.
Biophys.J., 102, 2012
|
|
4BBT
 
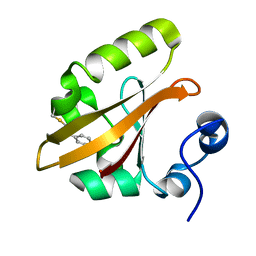 | | The PR1 Photocycle Intermediate of Photoactive Yellow Protein | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Schotte, F, Cho, H.S, Kaila, V.R.I, Kamikubo, H, Dashdorj, N, Henry, E.R, Graber, T.J, Henning, R, Wulff, M, Hummer, G, Kataoka, M, Anfinrud, P.A. | | Deposit date: | 2012-09-27 | | Release date: | 2012-11-14 | | Last modified: | 2019-01-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Watching a Signaling Protein Function in Real Time Via 100-Ps Time-Resolved Laue Crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4B9O
 
 | | The PR0 Photocycle Intermediate of Photoactive Yellow Protein | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Schotte, F, Cho, H.S, Kaila, V.R.I, Kamikubo, H, Dashdorj, N, Henry, E.R, Graber, T.J, Henning, R, Wulff, M, Hummer, G, Kataoka, M, Anfinrud, P.A. | | Deposit date: | 2012-09-06 | | Release date: | 2012-11-14 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Watching a Signaling Protein Function in Real Time Via 100-Ps Time-Resolved Laue Crystallography
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4BBV
 
 | | The PB0 Photocycle Intermediate of Photoactive Yellow Protein | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Schotte, F, Cho, H.S, Kaila, V.R.I, Kamikubo, H, Dashdorj, N, Henry, E.R, Graber, T.J, Henning, R, Wulff, M, Hummer, G, Kataoka, M, Anfinrud, P.A. | | Deposit date: | 2012-09-28 | | Release date: | 2012-11-14 | | Last modified: | 2019-01-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Watching a Signaling Protein Function in Real Time Via 100-Ps Time-Resolved Laue Crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4BBU
 
 | | The PR2 Photocycle Intermediate of Photoactive Yellow Protein | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Schotte, F, Cho, H.S, Kaila, V.R.I, Kamikubo, H, Dashdorj, N, Henry, E.R, Graber, T.J, Henning, R, Wulff, M, Hummer, G, Kataoka, M, Anfinrud, P.A. | | Deposit date: | 2012-09-27 | | Release date: | 2012-11-14 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Watching a Signaling Protein Function in Real Time Via 100-Ps Time-Resolved Laue Crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4WLA
 
 | |
4WL9
 
 | |
5HD3
 
 | |
5HDC
 
 | |
