5DWZ
 
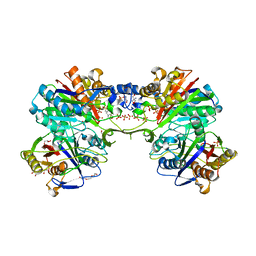 | | Structural and functional characterization of PqsBC, a condensing enzyme in the biosynthesis of the Pseudomonas aeruginosa quinolone signal | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-oxoacyl-(Acyl carrier protein) synthase III, 3-oxoacyl-[acyl-carrier-protein] synthase 3, ... | | Authors: | Drees, S.L, Li, C, Prasetya, F, Saleem, M, Dreveny, I, Hennecke, U, Williams, P, Emsley, J, Fetzner, S. | | Deposit date: | 2015-09-23 | | Release date: | 2016-02-03 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | PqsBC, a Condensing Enzyme in the Biosynthesis of the Pseudomonas aeruginosa Quinolone Signal: CRYSTAL STRUCTURE, INHIBITION, AND REACTION MECHANISM.
J.Biol.Chem., 291, 2016
|
|
2WB2
 
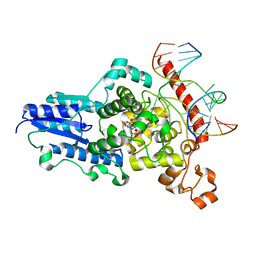 | | Drosophila Melanogaster (6-4) Photolyase Bound To double stranded Dna containing a T(6-4)C Photolesion | | Descriptor: | 5'-D(*AP*CP*AP*GP*CP*GP*GP*64PP*ZP*GP*CP*AP *GP*GP*T)-3', 5'-D(*TP*AP*CP*CP*TP*GP*CP*GP*AP*CP*CP*GP*CP*TP*G)-3', FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Glas, A.F, Schneider, S, Maul, M.J, Hennecke, U, Carell, T. | | Deposit date: | 2009-02-20 | | Release date: | 2009-03-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of the T(6-4)C Lesion in Complex with a (6-4) DNA Photolyase and Repair of Uv- Induced (6-4) and Dewar Photolesions.
Chemistry, 15, 2009
|
|
2VTB
 
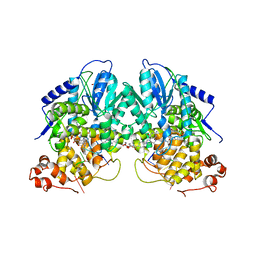 | | Structure of cryptochrome 3 - DNA complex | | Descriptor: | 5'-D(*DT*DT*DT*DT*DTP)-3', 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, ACETATE ION, ... | | Authors: | Pokorny, R, Klar, T, Hennecke, U, Carell, T, Batschauer, A, Essen, L.-O. | | Deposit date: | 2008-05-13 | | Release date: | 2009-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Recognition and Repair of Uv Lesions in Loop Structures of Duplex DNA by Dash-Type Cryptochrome.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6FHO
 
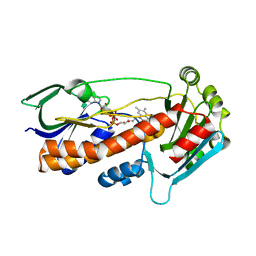 | | Crystal structure of pqsL, a probable FAD-dependent monooxygenase from Pseudomonas aeruginosa - new refinement | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable FAD-dependent monooxygenase | | Authors: | Belviso, B.D, Drees, S.L, Ernst, S, Jagmann, N, Hennecke, U, Fetzner, S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-04-25 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | PqsL uses reduced flavin to produce 2-hydroxylaminobenzoylacetate, a preferred PqsBC substrate in alkyl quinolone biosynthesis inPseudomonas aeruginosa.
J. Biol. Chem., 293, 2018
|
|
2J09
 
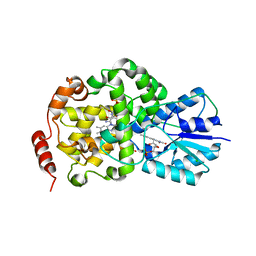 | | Thermus DNA photolyase with FMN antenna chromophore | | Descriptor: | CHLORIDE ION, DEOXYRIBODIPYRIMIDINE PHOTO-LYASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Klar, T, Kaiser, G, Hennecke, U, Carell, T, Batschauer, A, Essen, L.-O. | | Deposit date: | 2006-08-01 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Natural and Non-Natural Antenna Chromophores in the DNA Photolyase from Thermus Thermophilus
Chembiochem, 7, 2006
|
|
2J07
 
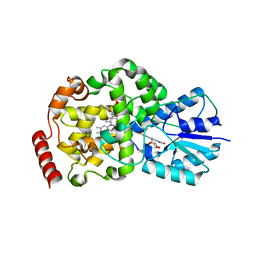 | | Thermus DNA photolyase with 8-HDF antenna chromophore | | Descriptor: | 8-HYDROXY-10-(D-RIBO-2,3,4,5-TETRAHYDROXYPENTYL)-5-DEAZAISOALLOXAZINE, CHLORIDE ION, DEOXYRIBODIPYRIMIDINE PHOTO-LYASE, ... | | Authors: | Klar, T, Kaiser, G, Hennecke, U, Carell, T, Batschauer, A, Essen, L.-O. | | Deposit date: | 2006-08-01 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Natural and Non-Natural Antenna Chromophores in the DNA Photolyase from Thermus Thermophilus
Chembiochem, 7, 2006
|
|
2J08
 
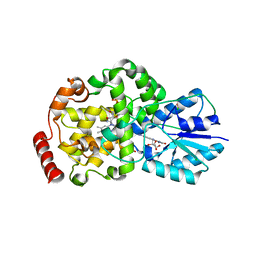 | | Thermus DNA photolyase with 8-Iod-riboflavin antenna chromophore | | Descriptor: | 1-DEOXY-1-(8-IODO-7-METHYL-2,4-DIOXO-3,4-DIHYDROBENZO[G]PTERIDIN-10(2H)-YL)-D-RIBITOL, CHLORIDE ION, DEOXYRIBODIPYRIMIDINE PHOTO-LYASE, ... | | Authors: | Klar, T, Kaiser, G, Hennecke, U, Carell, T, Batschauer, A, Essen, L.-O. | | Deposit date: | 2006-08-01 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Natural and Non-Natural Antenna Chromophores in the DNA Photolyase from Thermus Thermophilus
Chembiochem, 7, 2006
|
|
2JA5
 
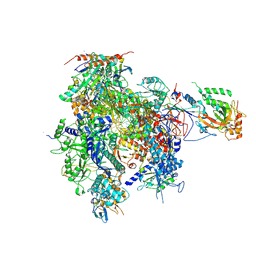 | | CPD lesion containing RNA Polymerase II elongation complex A | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP *CP*TP*TTP*TP*TP*CP*CP*BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*AP)-3', DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Brueckner, F, Hennecke, U, Carell, T, Cramer, P. | | Deposit date: | 2006-11-23 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Cpd Damage Recognition by Transcribing RNA Polymerase II
Science, 315, 2007
|
|
2JA8
 
 | | CPD lesion containing RNA Polymerase II elongation complex D | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP *CP*TP*TP*TP*TP*TTP*CP*BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-D(*TP*AP*AP*GP*TP*AP*CP*TP*TP*GP *AP*GP*CP*T)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*AP*UP)-3', ... | | Authors: | Brueckner, F, Hennecke, U, Carell, T, Cramer, P. | | Deposit date: | 2006-11-23 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Cpd Damage Recognition by Transcribing RNA Polymerase II.
Science, 315, 2007
|
|
2JA6
 
 | | CPD lesion containing RNA Polymerase II elongation complex B | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP *CP*TP*TP*TTP*TP*CP*CP*BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-D(*TP*AP*AP*GP*TP*AP*CP*TP*TP*GP *AP*GP*CP*T)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*AP)-3', ... | | Authors: | Brueckner, F, Hennecke, U, Carell, T, Cramer, P. | | Deposit date: | 2006-11-23 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | CPD damage recognition by transcribing RNA polymerase II.
Science, 315, 2007
|
|
2JA7
 
 | | CPD lesion containing RNA Polymerase II elongation complex C | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP *CP*TP*TP*TP*TTP*CP*CP*BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-D(*TP*AP*AP*GP*TP*AP*CP*TP*TP*GP *AP*GP*CP*T)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*AP)-3', ... | | Authors: | Brueckner, F, Hennecke, U, Carell, T, Cramer, P. | | Deposit date: | 2006-11-23 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Cpd Damage Recognition by Transcribing RNA Polymerase II.
Science, 315, 2007
|
|
1TEZ
 
 | | COMPLEX BETWEEN DNA AND THE DNA PHOTOLYASE FROM ANACYSTIS NIDULANS | | Descriptor: | 5'-D(*AP*TP*CP*GP*GP*CP*T*(TCP)P*CP*GP*C)-3', 5'-D(*TP*CP*GP*C)-3', 5'-D(P*CP*GP*AP*AP*GP*CP*CP*GP*A)-3', ... | | Authors: | Essen, L.-O, Carell, T, Mees, A, Klar, T. | | Deposit date: | 2004-05-26 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a photolyase bound to a CPD-like DNA lesion after in situ repair
Science, 306, 2004
|
|
6RB3
 
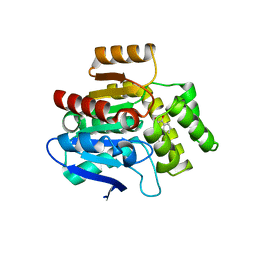 | | Structural basis for recognition and ring-cleavage of the Pseudomonas quinolone signal (PQS) by AqdC variant in complex with its substrate | | Descriptor: | 2-heptylquinoline-3,4-diol, Putative dioxygenase (1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase) | | Authors: | Wullich, S, Kobus, S, Smits, S.H, Fetzner, S. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recognition and ring-cleavage of the Pseudomonas quinolone signal (PQS) by AqdC, a mycobacterial dioxygenase of the alpha / beta-hydrolase fold family.
J.Struct.Biol., 207, 2019
|
|
6RA3
 
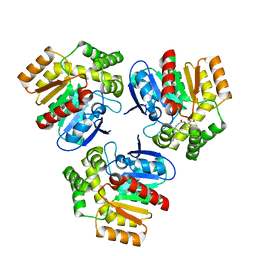 | | Structural basis for recognition and ring-cleavage of the Pseudomonas quinolone signal (PQS) by AqDC in complex with its product | | Descriptor: | 2-(octanoylamino)benzoic acid, Putative dioxygenase (1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase) | | Authors: | Wullich, S, Kobus, S, Smits, S.H, Fetzner, S. | | Deposit date: | 2019-04-05 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for recognition and ring-cleavage of the Pseudomonas quinolone signal (PQS) by AqdC, a mycobacterial dioxygenase of the alpha / beta-hydrolase fold family.
J.Struct.Biol., 207, 2019
|
|
6RA2
 
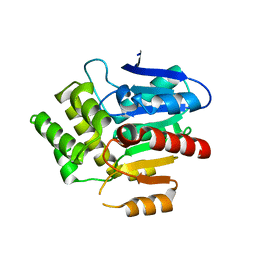 | | Structural basis for recognition and ring-cleavage of the Pseudomonas quinolone signal (PQS) by AqDC | | Descriptor: | Putative dioxygenase (1H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase) | | Authors: | Wullich, S, Kobus, S, Smits, S.H, Fetzner, S. | | Deposit date: | 2019-04-05 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recognition and ring-cleavage of the Pseudomonas quinolone signal (PQS) by AqdC, a mycobacterial dioxygenase of the alpha / beta-hydrolase fold family.
J.Struct.Biol., 207, 2019
|
|
