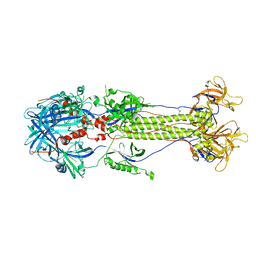
2GUM
 
 | |
1EXJ
 
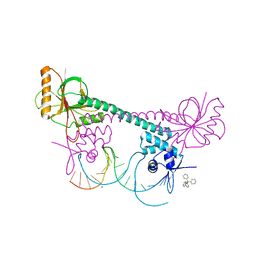 | | CRYSTAL STRUCTURE OF TRANSCRIPTION ACTIVATOR BMRR, FROM B. SUBTILIS, BOUND TO 21 BASE PAIR BMR OPERATOR AND TPP | | Descriptor: | DNA (5'-D(*AP*CP*CP*CP*TP*CP*CP*CP*CP*TP*TP*AP*GP*GP*GP*GP*AP*GP*GP*GP*T)-3'), MULTIDRUG-EFFLUX TRANSPORTER REGULATOR, SODIUM ION, ... | | Authors: | Zheleznova-Heldwein, E.E, Brennan, R.G. | | Deposit date: | 2000-05-02 | | Release date: | 2001-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the transcription activator BmrR bound to DNA and a drug.
Nature, 409, 2001
|
|
1EXI
 
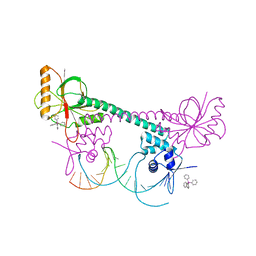 | | CRYSTAL STRUCTURE OF TRANSCRIPTION ACTIVATOR BMRR, FROM B. SUBTILIS, BOUND TO 21 BASE PAIR BMR OPERATOR AND TPSB | | Descriptor: | DNA (5'-D(*AP*CP*CP*CP*TP*CP*CP*CP*CP*TP*TP*AP*GP*GP*GP*GP*AP*GP*GP*GP*T)-3'), MULTIDRUG-EFFLUX TRANSPORTER REGULATOR, TETRAPHENYLANTIMONIUM ION, ... | | Authors: | Zheleznova-Heldwein, E.E, Brennan, R.G. | | Deposit date: | 2000-05-02 | | Release date: | 2001-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Crystal structure of the transcription activator BmrR bound to DNA and a drug.
Nature, 409, 2001
|
|
5J2Z
 
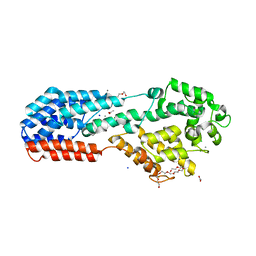 | | PRV UL37 N-terminal half (R2 mutant) | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Heldwein, E.E, Pitts, J.D. | | Deposit date: | 2016-03-30 | | Release date: | 2017-10-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The pUL37 tegument protein guides alpha-herpesvirus retrograde axonal transport to promote neuroinvasion.
PLoS Pathog., 13, 2017
|
|
4HSI
 
 | |
4U4H
 
 | |
7T7I
 
 | | EBV nuclear egress complex | | Descriptor: | Nuclear egress protein 1, Nuclear egress protein 2, ZINC ION | | Authors: | Thorsen, M.K, Heldwein, E.E. | | Deposit date: | 2021-12-15 | | Release date: | 2022-07-06 | | Last modified: | 2022-10-26 | | Method: | X-RAY DIFFRACTION (3.97 Å) | | Cite: | The nuclear egress complex of Epstein-Barr virus buds membranes through an oligomerization-driven mechanism.
Plos Pathog., 18, 2022
|
|
8G6D
 
 | |
6BM8
 
 | | Crystal structure of glycoprotein B from Herpes Simplex Virus type I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein B, ... | | Authors: | Cooper, R.S, Heldwein, E.E. | | Deposit date: | 2017-11-13 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural basis for membrane anchoring and fusion regulation of the herpes simplex virus fusogen gB.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5CXF
 
 | |
5ED7
 
 | | Crystal Structure of HSV-1 UL21 C-terminal Domain | | Descriptor: | CHLORIDE ION, Tegument protein UL21 | | Authors: | Metrick, C.M, Heldwein, E.E. | | Deposit date: | 2015-10-20 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Novel Structure and Unexpected RNA-Binding Ability of the C-Terminal Domain of Herpes Simplex Virus 1 Tegument Protein UL21.
J.Virol., 90, 2016
|
|
3M1C
 
 | |
5V2S
 
 | | Crystal structure of glycoprotein B from Herpes Simplex Virus type I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein B | | Authors: | Cooper, R.S, Heldwein, E.E. | | Deposit date: | 2017-03-06 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for membrane anchoring and fusion regulation of the herpes simplex virus fusogen gB.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5VYL
 
 | |
4K70
 
 | |
4Z3U
 
 | | PRV nuclear egress complex | | Descriptor: | CHLORIDE ION, SODIUM ION, UL31, ... | | Authors: | Bigalke, J.M, Heldwein, E.E. | | Deposit date: | 2015-03-31 | | Release date: | 2015-11-11 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | Structural basis of membrane budding by the nuclear egress complex of herpesviruses.
Embo J., 34, 2015
|
|
4ZXS
 
 | | HSV-1 nuclear egress complex | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, SODIUM ION, ... | | Authors: | Bigalke, J.M, Heldwein, E.E. | | Deposit date: | 2015-05-20 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.772 Å) | | Cite: | Structural basis of membrane budding by the nuclear egress complex of herpesviruses.
Embo J., 34, 2015
|
|
3NWA
 
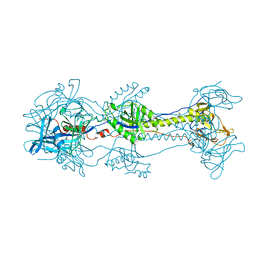 | | Glycoprotein B from Herpes simplex virus type 1, W174R mutant, low-pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein B, MESO-ERYTHRITOL | | Authors: | Stampfer, S.D, Lou, H, Cohen, G.H, Eisenberg, R.J, Heldwein, E.E. | | Deposit date: | 2010-07-09 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2629 Å) | | Cite: | Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1.
J.Virol., 84, 2010
|
|
3NWF
 
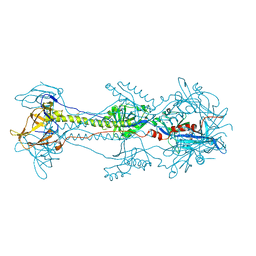 | | Glycoprotein B from Herpes simplex virus type 1, low-pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein B, MESO-ERYTHRITOL, ... | | Authors: | Stampfer, S.D, Lou, H, Cohen, G.H, Eisenberg, R.J, Heldwein, E.E. | | Deposit date: | 2010-07-09 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1.
J.Virol., 84, 2010
|
|
3NW8
 
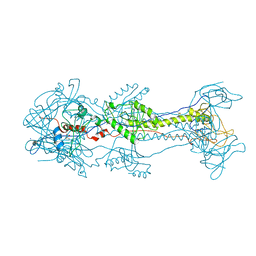 | | Glycoprotein B from Herpes simplex virus type 1, Y179S mutant, high-pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Stampfer, S.D, Lou, H, Cohen, G.H, Eisenberg, R.J, Heldwein, E.E. | | Deposit date: | 2010-07-09 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75915 Å) | | Cite: | Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1.
J.Virol., 84, 2010
|
|
3NWD
 
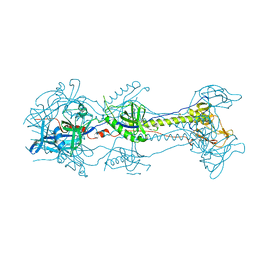 | | Glycoprotein B from Herpes simplex virus type 1, Y179S mutant, low-pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Stampfer, S.D, Lou, H, Cohen, G.H, Eisenberg, R.J, Heldwein, E.E. | | Deposit date: | 2010-07-09 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8803 Å) | | Cite: | Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1.
J.Virol., 84, 2010
|
|
1R8E
 
 | | Crystal Structure of BmrR Bound to DNA at 2.4A Resolution | | Descriptor: | 5'-D(*GP*AP*CP*CP*CP*TP*CP*CP*CP*CP*TP*TP*AP*GP*GP*GP*GP*AP*GP*GP*GP*TP*C)-3', GLYCEROL, IMIDAZOLE, ... | | Authors: | Newberry, K.J, Brennan, R.G. | | Deposit date: | 2003-10-23 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural mechanism for transcription activation by MerR family member multidrug transporter activation, N terminus.
J.Biol.Chem., 279, 2004
|
|
4L1R
 
 | |
