1RMJ
 
 | | C-terminal domain of insulin-like growth factor (IGF) binding protein-6: structure and interaction with IGF-II | | Descriptor: | Insulin-like growth factor binding protein 6 | | Authors: | Headey, S.J, Keizer, D.W, Yao, S, Brasier, G, Kantharidis, P, Bach, L.A, Norton, R.S. | | Deposit date: | 2003-11-28 | | Release date: | 2004-09-14 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | C-terminal domain of insulin-like growth factor (IGF) binding protein-6: structure and interaction with IGF-II.
Mol.Endocrinol., 18, 2004
|
|
2KXG
 
 | | The solution structure of the squash aspartic acid proteinase inhibitor (SQAPI) | | Descriptor: | Aspartic protease inhibitor | | Authors: | Headey, S.J, Macaskill, U.K, Wright, M, Claridge, J.K, Edwards, P.J.B, Farley, P.C, Christeller, J.T, Laing, W.A, Pascal, S.M. | | Deposit date: | 2010-05-05 | | Release date: | 2010-06-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the squash aspartic acid proteinase inhibitor (SQAPI) and mutational analysis of pepsin inhibition.
J.Biol.Chem., 285, 2010
|
|
2MHC
 
 | | NMR structure of the catalytic domain of the large serine resolvase TnpX | | Descriptor: | TnpX | | Authors: | Headey, S.J, Sivakumaran, A, Adams, V, Rodgers, A.J.W, Rood, J.I, Scanlon, M.J, Wilce, M.C.J. | | Deposit date: | 2013-11-20 | | Release date: | 2014-11-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and DNA Binding of the Catalytic of the Large Serine Resolvase Tnpx
To be Published
|
|
3OVN
 
 | | Fragment-based approach to the design of ligands targeting a novel site on HIV-1 integrase | | Descriptor: | 1-methyl-3-(thiophen-2-yl)-1H-pyrazol-5-amine, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Headey, S.J, Deadman, J.J, Rhodes, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2010-09-16 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment-based design of ligands targeting a novel site on the integrase enzyme of human immunodeficiency virus 1
Chemmedchem, 6, 2011
|
|
5TLQ
 
 | | Model structure of the oxidized PaDsbA1 and 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine complex | | Descriptor: | 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine, Thiol:disulfide interchange protein DsbA | | Authors: | Mohanty, B, Rimmer, K.A, McMahon, R.M, Headey, S.J, Vazirani, M, Shouldice, S.R, Coincon, M, Tay, S, Morton, C.J, Simpson, J.S, Martin, J.L, Scanlon, M.S. | | Deposit date: | 2016-10-11 | | Release date: | 2017-04-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Fragment library screening identifies hits that bind to the non-catalytic surface of Pseudomonas aeruginosa DsbA1.
PLoS ONE, 12, 2017
|
|
4WF5
 
 | | Crystal structure of E.Coli DsbA soaked with compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-2-[4-(trifluoromethyl)phenyl]-1,3-thiazole-5-carboxylic acid, COPPER (II) ION, ... | | Authors: | Adams, L.A, Sharma, P, Mohanty, B, Ilyichova, O.V, Mulcair, M.D, Williams, M.L, Gleeson, E.C, Totsika, M, Doak, B.C, Caria, S, Rimmer, K, Shouldice, S.R, Vazirani, M, Headey, S.J, Plumb, B.R, Martin, J.L, Heras, B, Simpson, J.S, Scanlon, M.J. | | Deposit date: | 2014-09-12 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Application of Fragment-Based Screening to the Design of Inhibitors of Escherichia coli DsbA.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4WEY
 
 | | Crystal structure of E.Coli DsbA in complex with compound 17 | | Descriptor: | 1,2-ETHANEDIOL, N-({4-methyl-2-[4-(trifluoromethyl)phenyl]-1,3-thiazol-5-yl}carbonyl)-L-serine, Thiol:disulfide interchange protein | | Authors: | Adams, L.A, Sharma, P, Mohanty, B, Ilyichova, O.V, Mulcair, M.D, Williams, M.L, Gleeson, E.C, Totsika, M, Doak, B.C, Caria, S, Rimmer, K, Shouldice, S.R, Vazirani, M, Headey, S.J, Plumb, B.R, Martin, J.L, Heras, B, Simpson, J.S, Scanlon, M.J. | | Deposit date: | 2014-09-11 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Application of Fragment-Based Screening to the Design of Inhibitors of Escherichia coli DsbA.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4WF4
 
 | | Crystal structure of E.Coli DsbA co-crystallised in complex with compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-2-[4-(trifluoromethyl)phenyl]-1,3-thiazole-5-carboxylic acid, Thiol:disulfide interchange protein | | Authors: | Adams, L.A, Sharma, P, Mohanty, B, Ilyichova, O.V, Mulcair, M.D, Williams, M.L, Gleeson, E.C, Totsika, M, Doak, B.C, Caria, S, Rimmer, K, Shouldice, S.R, Vazirani, M, Headey, S.J, Plumb, B.R, Martin, J.L, Heras, B, Simpson, J.S, Scanlon, M.J. | | Deposit date: | 2014-09-12 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Application of Fragment-Based Screening to the Design of Inhibitors of Escherichia coli DsbA.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3AO2
 
 | | Fragment-based approach to the design of ligands targeting a novel site on HIV-1 integrase | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3-(7-bromo-1,3-benzodioxol-5-yl)-1-methyl-1H-pyrazol-5-amine, ... | | Authors: | Wielens, J, Chalmers, D.K, Headey, S.J, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2010-09-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-based design of ligands targeting a novel site on the integrase enzyme of human immunodeficiency virus 1
Chemmedchem, 6, 2011
|
|
3AO5
 
 | | Fragment-based approach to the design of ligands targeting a novel site on HIV-1 integrase | | Descriptor: | 5-(7-bromo-1,3-benzodioxol-5-yl)-1-methyl-1H-pyrazol-3-amine, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Wielens, J, Headey, S.J, Parker, M.W, Chalmers, D.K, Scanlon, M.J. | | Deposit date: | 2010-09-21 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-based design of ligands targeting a novel site on the integrase enzyme of human immunodeficiency virus 1
Chemmedchem, 6, 2011
|
|
3AO4
 
 | | Fragment-based approach to the design of ligands targeting a novel site on HIV-1 integrase | | Descriptor: | 3-(1,3-benzodioxol-5-yl)-1-methyl-1H-pyrazol-5-amine, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Wielens, J, Headey, S.J, Parker, M.W, Chalmers, D.K, Scanlon, M.J. | | Deposit date: | 2010-09-20 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment-based design of ligands targeting a novel site on the integrase enzyme of human immunodeficiency virus 1
Chemmedchem, 6, 2011
|
|
3AO3
 
 | | Fragment-based approach to the design of ligands targeting a novel site on HIV-1 integrase | | Descriptor: | 3-(1,3-benzodioxol-5-yl)-1-methyl-1H-pyrazole-5-carboxylic acid, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Wielens, J, Headey, S.J, Parker, M.W, Chalmers, D.K, Scanlon, M.J. | | Deposit date: | 2010-09-20 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-based design of ligands targeting a novel site on the integrase enzyme of human immunodeficiency virus 1
Chemmedchem, 6, 2011
|
|
5DCH
 
 | | Crystal structure of Pseudomonas aeruginosa DsbA E82I in complex with MIPS-0000851 (3-[(2-METHYLBENZYL)SULFANYL]-4H-1,2,4-TRIAZOL-4-AMINE) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine, GLYCEROL, ... | | Authors: | McMahon, R.M, Martin, J.L. | | Deposit date: | 2015-08-24 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.447 Å) | | Cite: | Fragment library screening identifies hits that bind to the non-catalytic surface of Pseudomonas aeruginosa DsbA1.
PLoS ONE, 12, 2017
|
|
3L3V
 
 | | Structure of HIV-1 integrase core domain in complex with sucrose | | Descriptor: | CADMIUM ION, POL polyprotein, SULFATE ION, ... | | Authors: | Wielens, J, Chalmers, D.K, Scanlon, M.J, Parker, M.W. | | Deposit date: | 2009-12-18 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the HIV-1 integrase core domain in complex with sucrose reveals details of an allosteric inhibitory binding site
Febs Lett., 584, 2010
|
|
3L3U
 
 | | Crystal structure of the HIV-1 integrase core domain to 1.4A | | Descriptor: | POL polyprotein, SULFATE ION | | Authors: | Wielens, J, Chalmers, D.K, Scanlon, M.J, Parker, M.W. | | Deposit date: | 2009-12-17 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the HIV-1 integrase core domain in complex with sucrose reveals details of an allosteric inhibitory binding site.
Febs Lett., 584, 2010
|
|
4WET
 
 | | Crystal structure of E.Coli DsbA in complex with compound 16 | | Descriptor: | 1,2-ETHANEDIOL, N-({4-methyl-2-[4-(trifluoromethyl)phenyl]-1,3-thiazol-5-yl}carbonyl)-L-tyrosine, SODIUM ION, ... | | Authors: | Ilyichova, O.V, Scanlon, M.J. | | Deposit date: | 2014-09-11 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Application of Fragment-Based Screening to the Design of Inhibitors of Escherichia coli DsbA.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4Y1C
 
 | |
4Y1D
 
 | |
4AHV
 
 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-(1H-pyrazol-1-yl)phenyl]methanamine, ACETIC ACID, ... | | Authors: | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | Deposit date: | 2012-02-07 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
4AHR
 
 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | Descriptor: | 3-(1,3-benzodioxol-5-yl)propanoic acid, ACETIC ACID, GLYCEROL, ... | | Authors: | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | Deposit date: | 2012-02-07 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
4AH9
 
 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-PHENYL-1,2,4-THIADIAZOL-5-YL)-1,4-DIAZEPANE, CHLORIDE ION, ... | | Authors: | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | Deposit date: | 2012-02-06 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
4AHS
 
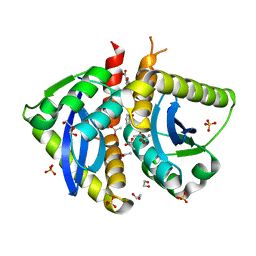 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | Descriptor: | 1,2-ETHANEDIOL, 1-BENZOFURAN-7-CARBOXYLIC ACID, ACETATE ION, ... | | Authors: | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | Deposit date: | 2012-02-07 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
2MJI
 
 | | HIFABP_Ketorolac_complex | | Descriptor: | (1R)-5-benzoyl-2,3-dihydro-1H-pyrrolizine-1-carboxylic acid, Fatty acid-binding protein, intestinal | | Authors: | Patil, R, Laguerre, A, Wielens, J, Headey, S, Williams, M, Mohanty, B, Porter, C, Scanlon, M. | | Deposit date: | 2014-01-09 | | Release date: | 2014-10-29 | | Last modified: | 2014-12-24 | | Method: | SOLUTION NMR | | Cite: | Characterization of two distinct modes of drug binding to human intestinal Fatty Acid binding protein.
Acs Chem.Biol., 9, 2014
|
|
2MIM
 
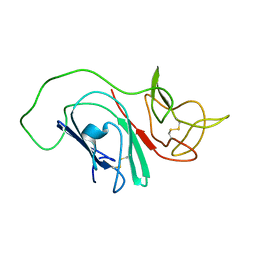 | |
2M32
 
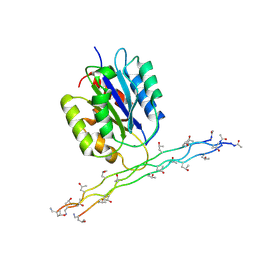 | | Alpha-1 integrin I-domain in complex with GLOGEN triple helical peptide | | Descriptor: | GLOGEN peptide, Integrin alpha-1, MAGNESIUM ION | | Authors: | Chin, Y, Headey, S, Mohanty, B, McEwan, P, Swarbrick, J, Mulhern, T, Emsley, J, Simpson, J, Scanlon, M. | | Deposit date: | 2013-01-07 | | Release date: | 2013-11-06 | | Last modified: | 2014-02-12 | | Method: | SOLUTION NMR | | Cite: | The Structure of Integrin alpha 1I Domain in Complex with a Collagen-mimetic Peptide.
J.Biol.Chem., 288, 2013
|
|
