1DGI
 
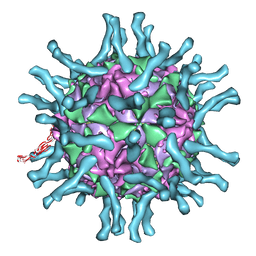 | | Cryo-EM structure of human poliovirus(serotype 1)complexed with three domain CD155 | | Descriptor: | POLIOVIRUS RECEPTOR, VP1, VP2, ... | | Authors: | He, Y, Bowman, V.D, Mueller, S, Bator, C.M, Bella, J, Peng, X, Baker, T.S, Wimmer, E, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 1999-11-24 | | Release date: | 2000-01-24 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (22 Å) | | Cite: | Interaction of the poliovirus receptor with poliovirus.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
6YXZ
 
 | |
6Z14
 
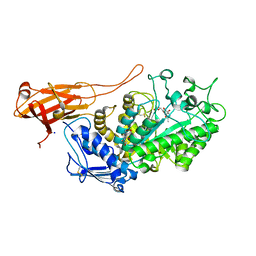 | | Structure of Bifidobacterium bifidum GH20 beta-N-beta-N-acetylhexosaminidase E553Q variant in complex with 4MU-6SGlcNAc-derived oxazoline | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-N-acetylhexosaminidase, NITRATE ION, ... | | Authors: | He, Y, Jin, Y, Rizkallah, P, Chen, P. | | Deposit date: | 2020-05-12 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure and activity of the GH20 beta-N-beta-N-acetylhexosaminidase from Bifidobacterium bifidum
To Be Published
|
|
6PNS
 
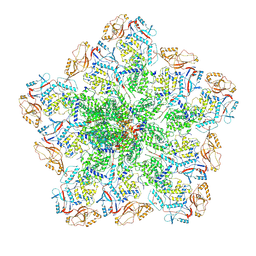 | | In situ structure of BTV RNA-dependent RNA polymerase in BTV virion | | Descriptor: | Inner core structural protein VP3, RNA-directed RNA polymerase | | Authors: | He, Y, Shivakoti, S, Ding, K, Cui, Y, Roy, P, Zhou, Z.H. | | Deposit date: | 2019-07-03 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | In situ structures of RNA-dependent RNA polymerase inside bluetongue virus before and after uncoating.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6PO2
 
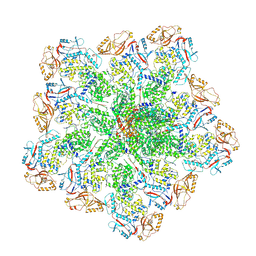 | | In situ structure of BTV RNA-dependent RNA polymerase in BTV core | | Descriptor: | Inner core structural protein VP3, RNA-directed RNA polymerase | | Authors: | He, Y, Shivakoti, S, Ding, K, Cui, Y, Roy, P, Zhou, Z.H. | | Deposit date: | 2019-07-03 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | In situ structures of RNA-dependent RNA polymerase inside bluetongue virus before and after uncoating.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7LMA
 
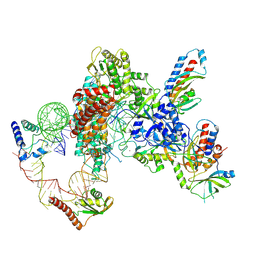 | | Tetrahymena telomerase T3D2 structure at 3.3 Angstrom | | Descriptor: | Telomerase La-related protein p65, Telomerase RNA, Telomerase associated protein p50, ... | | Authors: | He, Y, Wang, Y, Liu, B, Helmling, C, Susac, L, Cheng, R, Zhou, Z.H, Feigon, J. | | Deposit date: | 2021-02-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of telomerase at several steps of telomere repeat synthesis.
Nature, 593, 2021
|
|
7LMB
 
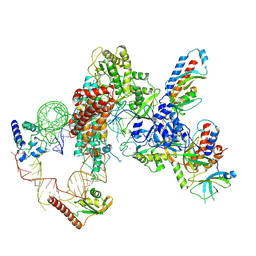 | | Tetrahymena telomerase T5D5 structure at 3.8 Angstrom | | Descriptor: | Telomerase La-related protein p65, Telomerase RNA, Telomerase associated protein p50, ... | | Authors: | He, Y, Wang, Y, Liu, B, Helmling, C, Susac, L, Cheng, R, Zhou, Z.H, Feigon, J. | | Deposit date: | 2021-02-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of telomerase at several steps of telomere repeat synthesis.
Nature, 593, 2021
|
|
2FS1
 
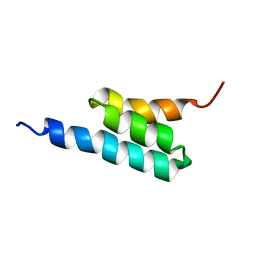 | | solution structure of PSD-1 | | Descriptor: | PSD-1 | | Authors: | He, Y, Rozak, D.A, Sari, N, Chen, Y, Bryan, P, Orban, J. | | Deposit date: | 2006-01-20 | | Release date: | 2006-12-05 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics, and stability variation in bacterial albumin binding modules: implications for species specificity.
Biochemistry, 45, 2006
|
|
3SQJ
 
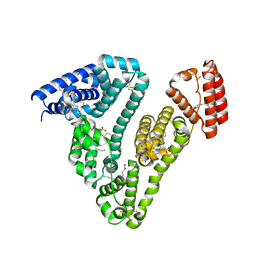 | | Recombinant human serum albumin from transgenic plant | | Descriptor: | MYRISTIC ACID, Serum albumin | | Authors: | He, Y, Yang, D. | | Deposit date: | 2011-07-05 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Large-scale production of functional human serum albumin from transgenic rice seeds.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1NN8
 
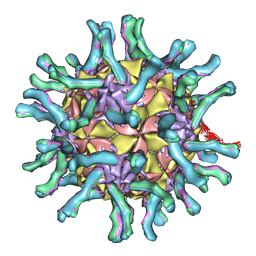 | | CryoEM structure of poliovirus receptor bound to poliovirus | | Descriptor: | MYRISTIC ACID, coat protein VP1, coat protein VP2, ... | | Authors: | He, Y, Mueller, S, Chipman, P.R, Bator, C.M, Peng, X, Bowman, V.D, Mukhopadhyay, S, Wimmer, E, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2003-01-13 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Complexes of poliovirus serotypes with their common cellular receptor, CD155
J.Virol., 77, 2003
|
|
4BMH
 
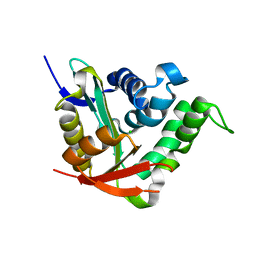 | | Crystal structure of SsHAT | | Descriptor: | ACETYLTRANSFERASE, CHLORIDE ION | | Authors: | He, Y, Turkenburg, J.P, Davies, G.J. | | Deposit date: | 2013-05-08 | | Release date: | 2014-01-15 | | Last modified: | 2014-01-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-Dimensional Structure of a Streptomyces Sviceus Gnat Acetyltransferase with Similarity to the C-Terminal Domain of the Human Gh84 O-Glcnacase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2GA5
 
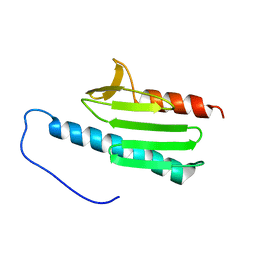 | | yeast frataxin | | Descriptor: | Frataxin homolog, mitochondrial | | Authors: | He, Y, Alam, S.L, Proteasa, S.V, Zhang, Y, Lesuisse, E, Dancis, A. | | Deposit date: | 2006-03-07 | | Release date: | 2006-03-21 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Yeast Frataxin Solution Structure, Iron Binding and Ferrochelatase Interaction
Biochemistry, 43, 2004
|
|
2JWS
 
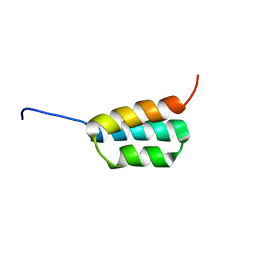 | | Solution NMR structures of two designed proteins with 88% sequence identity but different fold and function | | Descriptor: | Ga88 | | Authors: | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | Deposit date: | 2007-10-24 | | Release date: | 2008-09-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structures of two designed proteins with high sequence identity but different fold and function
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2JWU
 
 | | Solution NMR structures of two designed proteins with 88% sequence identity but different fold and function | | Descriptor: | Gb88 | | Authors: | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | Deposit date: | 2007-10-25 | | Release date: | 2008-09-09 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | NMR structures of two designed proteins with high sequence identity but different fold and function
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2KQ8
 
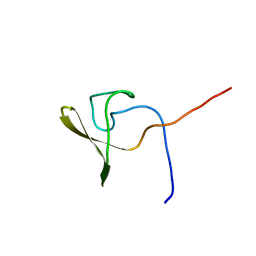 | | Solution NMR structure of a domain from BT9727_4915 from Bacillus thuringiensis, Northeast Structural Genomics Consortium Target BuR95A | | Descriptor: | Cell wall hydrolase | | Authors: | He, Y, Mills, J.L, Wu, Y, Eletsky, A, Wang, H, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-30 | | Release date: | 2010-01-26 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a domain from BT9727_4915 from Bacillus thuringiensis, Northeast Structural Genomics Consortium Target BuR95A
To be Published
|
|
2LHE
 
 | | Gb98-T25I,L20A | | Descriptor: | Gb98 | | Authors: | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | Deposit date: | 2011-08-08 | | Release date: | 2012-02-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Mutational tipping points for switching protein folds and functions.
Structure, 20, 2012
|
|
2LHG
 
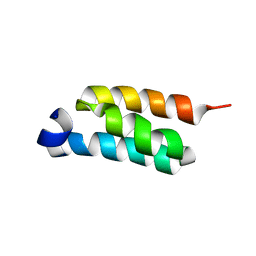 | | GB98-T25I solution structure | | Descriptor: | GB98 | | Authors: | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | Deposit date: | 2011-08-08 | | Release date: | 2012-02-29 | | Method: | SOLUTION NMR | | Cite: | Mutational tipping points for switching protein folds and functions.
Structure, 20, 2012
|
|
2LHC
 
 | | Ga98 solution structure | | Descriptor: | Ga98 | | Authors: | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | Deposit date: | 2011-08-08 | | Release date: | 2012-02-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Mutational tipping points for switching protein folds and functions.
Structure, 20, 2012
|
|
2LHD
 
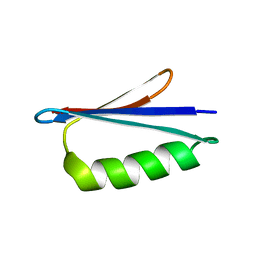 | | GB98 solution structure | | Descriptor: | GB98 | | Authors: | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | Deposit date: | 2011-08-08 | | Release date: | 2012-02-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Mutational tipping points for switching protein folds and functions.
Structure, 20, 2012
|
|
2LU1
 
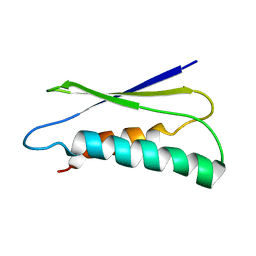 | | pfsub2 solution NMR structure | | Descriptor: | Subtilase | | Authors: | He, Y, Chen, Y, Ruan, B, O'Brochta, D, Bryan, P, Orban, J. | | Deposit date: | 2012-06-06 | | Release date: | 2012-10-03 | | Last modified: | 2012-11-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a sheddase inhibitor prodomain from the malarial parasite Plasmodium falciparum.
Proteins, 80, 2012
|
|
2WZH
 
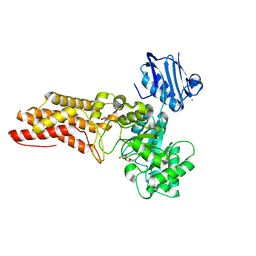 | | BtGH84 D242N in complex with MeUMB-derived oxazoline | | Descriptor: | 2-METHYL-4,5-DIHYDRO-(1,2-DIDEOXY-ALPHA-D-GLUCOPYRANOSO)[2,1-D]-1,3-OXAZOLE, CALCIUM ION, GLYCEROL, ... | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2009-11-30 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Visualizing the Reaction Coordinate of an O-Glcnac Hydrolase
J.Am.Chem.Soc., 132, 2010
|
|
2X0H
 
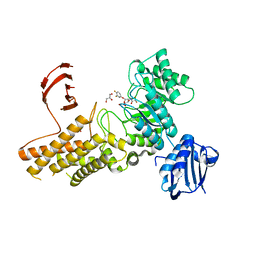 | | BtGH84 Michaelis complex | | Descriptor: | 3,4-difluorophenyl 2-deoxy-2-[(difluoroacetyl)amino]-beta-D-glucopyranoside, CALCIUM ION, GLYCEROL, ... | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2009-12-08 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Visualizing the Reaction Coordinate of an O-Glcnac Hydrolase
J.Am.Chem.Soc., 132, 2010
|
|
2WZI
 
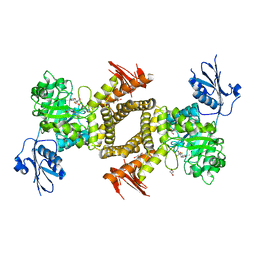 | | BtGH84 D243N in complex with 5F-oxazoline | | Descriptor: | (3AS,5S,6S,7R,7AR)-5-FLUORO-5-(HYDROXYMETHYL)-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]OXAZOLE-6,7-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2009-11-30 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Visualizing the Reaction Coordinate of an O-Glcnac Hydrolase
J.Am.Chem.Soc., 132, 2010
|
|
5I8Q
 
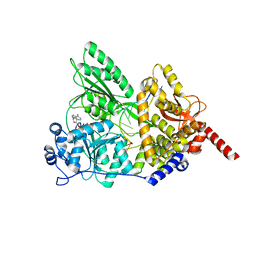 | | S. cerevisiae Prp43 in complex with RNA and ADPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP43, ... | | Authors: | He, Y, Nielsen, K.H, Andersen, G.R. | | Deposit date: | 2016-02-19 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structure of the DEAH/RHA ATPase Prp43p bound to RNA implicates a pair of hairpins and motif Va in translocation along RNA.
RNA, 23, 2017
|
|
5IY8
 
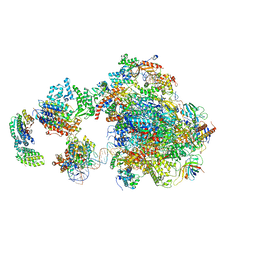 | | Human holo-PIC in the initial transcribing state | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | Deposit date: | 2016-03-24 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
