1K8D
 
 | | crystal structure of the non-classical MHC class Ib Qa-2 complexed with a self peptide | | Descriptor: | 60S RIBOSOMAL PROTEIN, BETA-2-MICROGLOBULIN, QA-2 antigen | | Authors: | He, X, Tabaczewski, P, Ho, J, Stroynowski, I, Garcia, K.C. | | Deposit date: | 2001-10-23 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Promiscuous antigen presentation by the nonclassical MHC Ib Qa-2 is enabled by a shallow, hydrophobic groove and self-stabilized peptide conformation.
Structure, 9, 2001
|
|
3CM8
 
 | | A RNA polymerase subunit structure from virus | | Descriptor: | Polymerase acidic protein, peptide from RNA-directed RNA polymerase catalytic subunit | | Authors: | He, X, Zhou, J, Zeng, Z, Ma, J, Zhang, R, Rao, Z, Liu, Y. | | Deposit date: | 2008-03-21 | | Release date: | 2008-07-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Crystal structure of the polymerase PAC-PB1N complex from an avian influenza H5N1 virus
Nature, 454, 2008
|
|
1Q32
 
 | | Crystal Structure Analysis of the Yeast Tyrosyl-DNA Phosphodiesterase | | Descriptor: | tyrosyl-DNA phosphodiesterase | | Authors: | He, X, Babaoglu, K, Price, A, Nitiss, K.C, Nitiss, J.L, White, S.W. | | Deposit date: | 2003-07-28 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mutation of a conserved active site residue converts tyrosyl-DNA phosphodiesterase I into a DNA topoisomerase I-dependent poison
J.Mol.Biol., 372, 2007
|
|
4TRJ
 
 | | Crystal structure of Mycobacterium tuberculosis enoyl reductase (INHA) complexed with N-(3-bromophenyl)-1-cyclohexyl-5-oxopyrrolidine-3-carboxamide, refined with new ligand restraints | | Descriptor: | (3S)-N-(3-BROMOPHENYL)-1-CYCLOHEXYL-5-OXOPYRROLIDINE-3-CARBOXAMIDE, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | He, X, Alian, A, Stroud, R.M, Ortiz de Montellano, P.R. | | Deposit date: | 2014-06-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Pyrrolidine carboxamides as a novel class of inhibitors of enoyl acyl carrier protein reductase from Mycobacterium tuberculosis
J. Med. Chem., 2006
|
|
4TZK
 
 | | Crystal structure of Mycobacterium tuberculosis enoyl reductase (INHA) complexed WITH 1-CYCLOHEXYL-N-(3,5-DICHLOROPHENYL)-5-OXOPYRROLIDINE-3-CARBOXAMIDE | | Descriptor: | (3S)-1-CYCLOHEXYL-N-(3,5-DICHLOROPHENYL)-5-OXOPYRROLIDINE-3-CARBOXAMIDE, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | He, X, Alian, A, Stroud, R.M, Ortiz de Montellano, P.R. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Pyrrolidine carboxamides as a novel class of inhibitors of enoyl acyl carrier protein reductase from Mycobacterium tuberculosis
J. Med. Chem., 2006
|
|
4U0J
 
 | | Crystal structure of Mycobacterium tuberculosis enoyl reductase (INHA) complexed with 1-CYCLOHEXYL-5-OXO-N-PHENYLPYRROLIDINE-3-CARBOXAMIDE, refined with new ligand restraints | | Descriptor: | (3S)-1-CYCLOHEXYL-5-OXO-N-PHENYLPYRROLIDINE-3-CARBOXAMIDE, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | He, X, Alian, A, Stroud, R.M, Ortiz de Montellano, P.R. | | Deposit date: | 2014-07-11 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Pyrrolidine carboxamides as a novel class of inhibitors of enoyl acyl carrier protein reductase from Mycobacterium tuberculosis
J. Med. Chem., 2006
|
|
4U0K
 
 | | Crystal structure of Mycobacterium tuberculosis enoyl reductase complexed with N-(5-chloro-2-methylphenyl)-1-cyclohexyl-5-oxopyrrolidine-3-carboxamide | | Descriptor: | (3S)-N-(5-CHLORO-2-METHYLPHENYL)-1-CYCLOHEXYL-5-OXOPYRROLIDINE-3-CARBOXAMIDE, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | He, X, Alian, A, Stroud, R.M, Ortiz de Montellano, P.R. | | Deposit date: | 2014-07-11 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyrrolidine carboxamides as a novel class of inhibitors of enoyl acyl carrier protein reductase from Mycobacterium tuberculosis
J. Med. Chem., 2006
|
|
4TZT
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS ENOYL REDUCTASE (INHA) COMPLEXED WITH N-(3-CHLORO-2-METHYLPHENYL)-1-CYCLOHEXYL- 5-OXOPYRROLIDINE-3-CARBOXAMIDE | | Descriptor: | (3S)-N-(3-CHLORO-2-METHYLPHENYL)-1-CYCLOHEXYL-5-OXOPYRROLIDINE-3-CARBOXAMIDE, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | He, X, Alian, A, Stroud, R.M, Ortiz de Montellano, P.R. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Pyrrolidine carboxamides as a novel class of inhibitors of enoyl acyl carrier protein reductase from Mycobacterium tuberculosis
J. Med. Chem., 2006
|
|
3UPU
 
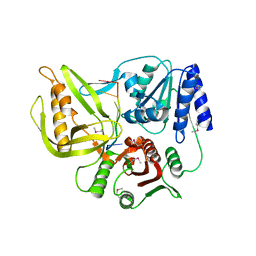 | | Crystal structure of the T4 Phage SF1B Helicase Dda | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*T)-3', ATP-dependent DNA helicase dda | | Authors: | He, X, Yun, M.K, Pemble IV, C.W, Kreuzer, K.N, Raney, K.D, White, S.W. | | Deposit date: | 2011-11-18 | | Release date: | 2012-06-20 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | The T4 Phage SF1B Helicase Dda Is Structurally Optimized to Perform DNA Strand Separation.
Structure, 20, 2012
|
|
4GYV
 
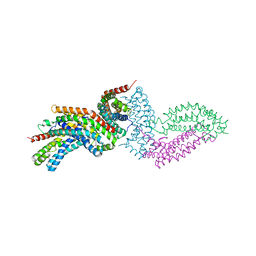 | | Crystal structure of the DH domain of FARP2 | | Descriptor: | FERM, RhoGEF and pleckstrin domain-containing protein 2 | | Authors: | He, X, Zhang, X. | | Deposit date: | 2012-09-05 | | Release date: | 2013-03-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for Autoinhibition of the Guanine Nucleotide Exchange Factor FARP2.
Structure, 21, 2013
|
|
4GZU
 
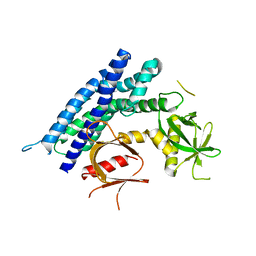 | | Crystal structure of the DH-PH-PH domain of FARP2 | | Descriptor: | FERM, RhoGEF and pleckstrin domain-containing protein 2 | | Authors: | He, X, Zhang, X. | | Deposit date: | 2012-09-06 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for Autoinhibition of the Guanine Nucleotide Exchange Factor FARP2.
Structure, 21, 2013
|
|
4H6Y
 
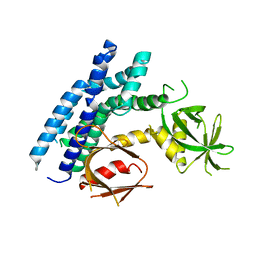 | | Crystal structure of the DH-PH-PH domain of FARP1 | | Descriptor: | FERM, RhoGEF and pleckstrin domain-containing protein 1 | | Authors: | He, X, Zhang, X. | | Deposit date: | 2012-09-19 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.09 Å) | | Cite: | Structural Basis for Autoinhibition of the Guanine Nucleotide Exchange Factor FARP2.
Structure, 21, 2013
|
|
2NSD
 
 | |
3MKT
 
 | | Structure of a Cation-bound Multidrug and Toxin Compound Extrusion (MATE) transporter | | Descriptor: | Multi antimicrobial extrusion protein (Na(+)/drug antiporter) MATE-like MDR efflux pump | | Authors: | He, X, Szewczyk, P, Karyakin, A, Evin, M, Hong, W.-X, Zhang, Q, Chang, G. | | Deposit date: | 2010-04-15 | | Release date: | 2010-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structure of a cation-bound multidrug and toxic compound extrusion transporter.
Nature, 467, 2010
|
|
3MKU
 
 | | Structure of a Cation-bound Multidrug and Toxin Compound Extrusion (MATE) transporter | | Descriptor: | Multi antimicrobial extrusion protein (Na(+)/drug antiporter) MATE-like MDR efflux pump, RUBIDIUM ION | | Authors: | He, X, Szewczyk, P, Karyakin, A, Evin, M, Hong, W.-X, Zhang, Q, Chang, G. | | Deposit date: | 2010-04-15 | | Release date: | 2010-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structure of a cation-bound multidrug and toxic compound extrusion transporter.
Nature, 467, 2010
|
|
8S9I
 
 | |
8GME
 
 | |
4FA8
 
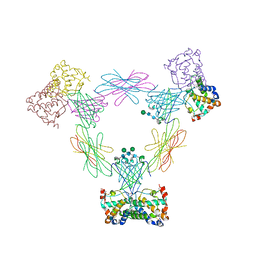 | | Multi-pronged modulation of cytokine signaling | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage colony-stimulating factor 1, Secreted protein BARF1, ... | | Authors: | He, X, Shim, A.H. | | Deposit date: | 2012-05-21 | | Release date: | 2012-08-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multipronged attenuation of macrophage-colony stimulating factor signaling by Epstein-Barr virus BARF1.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3HY5
 
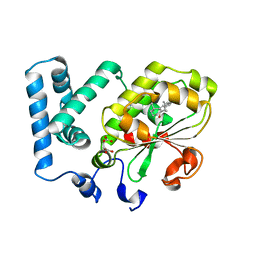 | | Crystal structure of CRALBP | | Descriptor: | L(+)-TARTARIC ACID, RETINAL, Retinaldehyde-binding protein 1 | | Authors: | Stocker, A, He, X, Lobsiger, J. | | Deposit date: | 2009-06-22 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Bothnia dystrophy is caused by domino-like rearrangements in cellular retinaldehyde-binding protein mutant R234W.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HX3
 
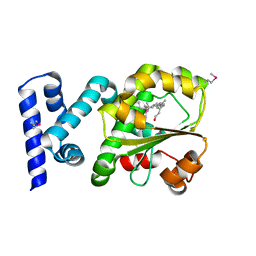 | | Crystal structure of CRALBP mutant R234W | | Descriptor: | RETINAL, Retinaldehyde-binding protein 1 | | Authors: | Stocker, A, He, X, Lobsiger, J. | | Deposit date: | 2009-06-19 | | Release date: | 2009-10-20 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Bothnia dystrophy is caused by domino-like rearrangements in cellular retinaldehyde-binding protein mutant R234W.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6D2K
 
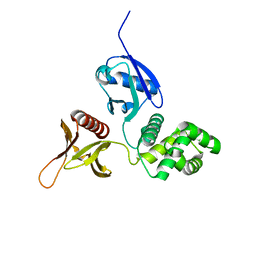 | | Crystal structure of the FERM domain of mouse FARP2 | | Descriptor: | FERM, ARHGEF and pleckstrin domain-containing protein 2 | | Authors: | He, X, Zhang, X. | | Deposit date: | 2018-04-13 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analyses of FERM domain-mediated membrane localization of FARP1.
Sci Rep, 8, 2018
|
|
2IFG
 
 | |
1S17
 
 | | Identification of Novel Potent Bicyclic Peptide Deformylase Inhibitors | | Descriptor: | 2-(3,4-DIHYDRO-3-OXO-2H-BENZO[B][1,4]THIAZIN-2-YL)-N-HYDROXYACETAMIDE, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Molteni, V, He, X, Nabakka, J, Yang, K, Kreusch, A, Gordon, P, Bursulaya, B, Ryder, N.S, Goldberg, R, He, Y. | | Deposit date: | 2004-01-05 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of novel potent bicyclic peptide deformylase inhibitors
Bioorg.Med.Chem.Lett., 14, 2004
|
|
2ACV
 
 | | Crystal Structure of Medicago truncatula UGT71G1 | | Descriptor: | URIDINE-5'-DIPHOSPHATE, triterpene UDP-glucosyl transferase UGT71G1 | | Authors: | Shao, H, He, X, Achnine, L, Blount, J.W, Dixon, R.A, Wang, X. | | Deposit date: | 2005-07-19 | | Release date: | 2005-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of a Multifunctional Triterpene/Flavonoid Glycosyltransferase from Medicago truncatula.
Plant Cell, 17, 2005
|
|
2ACW
 
 | | Crystal Structure of Medicago truncatula UGT71G1 complexed with UDP-glucose | | Descriptor: | URIDINE-5'-DIPHOSPHATE-GLUCOSE, triterpene UDP-glucosyl transferase UGT71G1 | | Authors: | Shao, H, He, X, Achnine, L, Blount, J.W, Dixon, R.A, Wang, X. | | Deposit date: | 2005-07-19 | | Release date: | 2005-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of a Multifunctional Triterpene/Flavonoid Glycosyltransferase from Medicago truncatula.
Plant Cell, 17, 2005
|
|
