5DA5
 
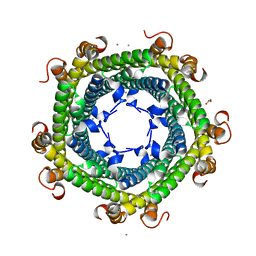 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 | | Descriptor: | CALCIUM ION, FE (III) ION, GLYCOLIC ACID, ... | | Authors: | He, D, Vanden Hehier, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2015-08-19 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.064 Å) | | Cite: | Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments.
Elife, 5, 2016
|
|
5L8B
 
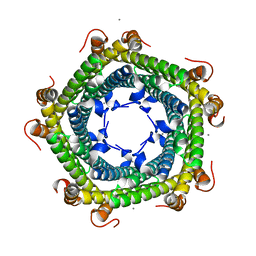 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant E62A | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2016-06-07 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments.
Elife, 5, 2016
|
|
5L8G
 
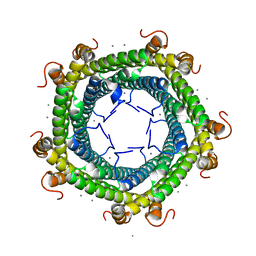 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant H65A | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2016-06-07 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.974 Å) | | Cite: | Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments.
Elife, 5, 2016
|
|
5L89
 
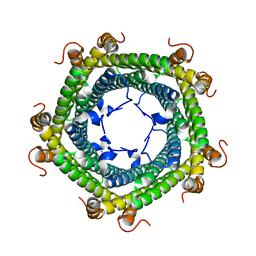 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant E32A | | Descriptor: | CALCIUM ION, Rru_A0973 | | Authors: | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2016-06-07 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments.
Elife, 5, 2016
|
|
5L8F
 
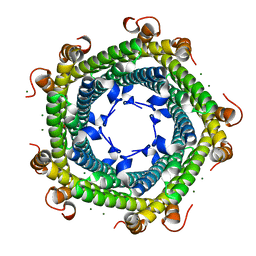 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant E32A, E62A, H65A. | | Descriptor: | MAGNESIUM ION, Rru_A0973, TRIETHYLENE GLYCOL | | Authors: | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2016-06-07 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant E32A, E62A, H65A.
To be published
|
|
5N5F
 
 | |
5N5E
 
 | |
2B3R
 
 | | Crystal structure of the C2 domain of class II phosphatidylinositide 3-kinase C2 | | Descriptor: | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide, SULFATE ION | | Authors: | Liu, L, Song, X, He, D, Komma, C, Kita, A, Verbasius, J.V, Bellamy, H, Miki, K, Czech, M.P, Zhou, G.W. | | Deposit date: | 2005-09-20 | | Release date: | 2005-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the C2 domain of class II phosphatidylinositide 3-kinase C2alpha.
J.Biol.Chem., 281, 2006
|
|
2B3O
 
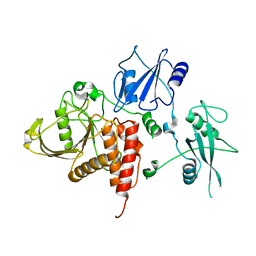 | | Crystal structure of human tyrosine phosphatase SHP-1 | | Descriptor: | Tyrosine-protein phosphatase, non-receptor type 6 | | Authors: | Yang, J, Liu, L, He, D, Song, X, Liang, X, Zhao, Z.J, Zhou, G.W. | | Deposit date: | 2005-09-20 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human protein-tyrosine phosphatase SHP-1.
J.Biol.Chem., 278, 2003
|
|
6SUW
 
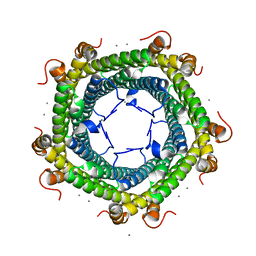 | |
6SV1
 
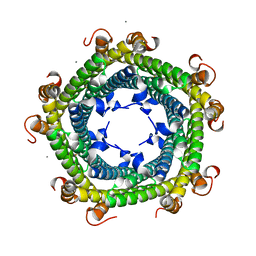 | |
5Z1V
 
 | | Crystal structure of AvrPib | | Descriptor: | AvrPib protein | | Authors: | Zhang, X, He, D, Zhao, Y.X, Taylor, I.A, Peng, Y.L, Yang, J, Liu, J.F. | | Deposit date: | 2017-12-28 | | Release date: | 2018-09-05 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (1.661 Å) | | Cite: | A positive-charged patch and stabilized hydrophobic core are essential for avirulence function of AvrPib in the rice blast fungus.
Plant J., 96, 2018
|
|
7F0R
 
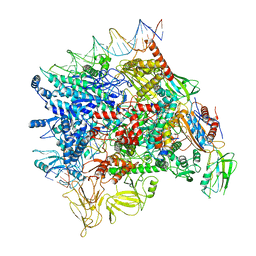 | | Cryo-EM structure of Pseudomonas aeruginosa SutA transcription activation complex | | Descriptor: | DNA (70-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2021-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7VF9
 
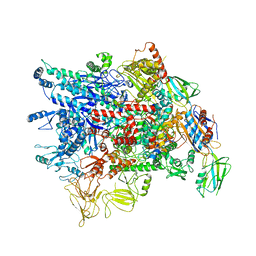 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2021-09-10 | | Release date: | 2022-07-27 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7XL3
 
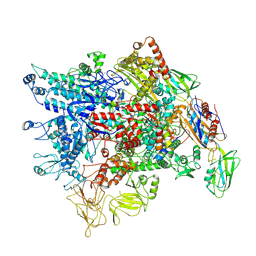 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes with transcription factor SutA (open lobe) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7XL4
 
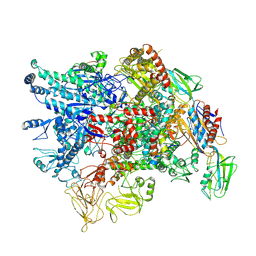 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes with transcription factor SutA (closed lobe) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-27 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
1O61
 
 | | Crystal structure of a PLP-dependent enzyme with PLP | | Descriptor: | ACETATE ION, PYRIDOXAL-5'-PHOSPHATE, aminotransferase | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O68
 
 | |
1O6D
 
 | | Crystal structure of a hypothetical protein | | Descriptor: | Hypothetical UPF0247 protein TM0844 | | Authors: | Structural GenomiX | | Deposit date: | 2003-11-03 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O69
 
 | | Crystal structure of a PLP-dependent enzyme | | Descriptor: | (2-AMINO-4-FORMYL-5-HYDROXY-6-METHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, BETA-MERCAPTOETHANOL, aminotransferase | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O64
 
 | |
1O62
 
 | |
1O65
 
 | | Crystal structure of an hypothetical protein | | Descriptor: | Hypothetical protein yiiM | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VHN
 
 | |
1VI7
 
 | | Crystal structure of an hypothetical protein | | Descriptor: | Hypothetical protein yigZ | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of YIGZ, a conserved hypothetical protein from Escherichia coli k12 with a novel fold
Proteins, 55, 2004
|
|
