1SVJ
 
 | | The solution structure of the nucleotide binding domain of KdpB | | Descriptor: | Potassium-transporting ATPase B chain | | Authors: | Haupt, M, Bramkamp, M, Coles, M, Altendorf, K, Kessler, H. | | Deposit date: | 2004-03-29 | | Release date: | 2004-09-21 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Inter-domain motions of the N-domain of the KdpFABC complex, a P-type ATPase, are not driven by ATP-induced conformational changes.
J.Mol.Biol., 342, 2004
|
|
1U7Q
 
 | | THE SOLUTION STRUCTURE OF THE NUCLEOTIDE BINDING DOMAIN OF KDPB | | Descriptor: | Potassium-transporting ATPase B chain | | Authors: | Haupt, M, Bramkamp, M, Coles, M, Altendorf, K, Kessler, H. | | Deposit date: | 2004-08-04 | | Release date: | 2004-09-21 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Inter-domain motions of the N-domain of the KdpFABC complex, a P-type ATPase, are not driven by ATP-induced conformational changes.
J.Mol.Biol., 342, 2004
|
|
2A29
 
 | | The solution structure of the AMP-PNP bound nucleotide binding domain of KdpB | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Potassium-transporting ATPase B chain | | Authors: | Haupt, M, Bramkamp, M, Coles, M, Altendorf, K, Kessler, H. | | Deposit date: | 2005-06-22 | | Release date: | 2005-12-20 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The Holo-form of the Nucleotide Binding Domain of the KdpFABC Complex from Escherichia coli Reveals a New Binding Mode
J.Biol.Chem., 281, 2006
|
|
2A00
 
 | | The solution structure of the AMP-PNP bound nucleotide binding domain of KdpB | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Potassium-transporting ATPase B chain | | Authors: | Haupt, M, Bramkamp, M, Coles, M, Altendorf, K, Kessler, H. | | Deposit date: | 2005-06-15 | | Release date: | 2005-12-20 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The Holo-form of the Nucleotide Binding Domain of the KdpFABC Complex from Escherichia coli Reveals a New Binding Mode
J.Biol.Chem., 281, 2006
|
|
4PVM
 
 | | Neutron structure of human transthyretin (TTR) at room temperature to 2.0A resolution (Laue) | | Descriptor: | Transthyretin | | Authors: | Fisher, S.J, Blakeley, M.P, Haupt, M, Mason, S.A, Cooper, J.B, Mitchell, E.P, Forsyth, V.T. | | Deposit date: | 2014-03-18 | | Release date: | 2014-11-12 | | Last modified: | 2024-03-20 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Binding site asymmetry in human transthyretin: insights from a joint neutron and X-ray crystallographic analysis using perdeuterated protein
IUCrJ, 1, 2014
|
|
4PVL
 
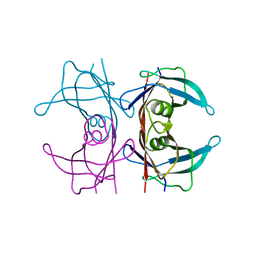 | | X-ray structure of human transthyretin (TTR) at room temperature to 1.9A resolution | | Descriptor: | Transthyretin | | Authors: | Fisher, S.J, Blakeley, M.P, Haupt, M, Mason, S.A, Cooper, J.B, Mitchell, E.P, Forsyth, V.T. | | Deposit date: | 2014-03-18 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Binding site asymmetry in human transthyretin: insights from a joint neutron and X-ray crystallographic analysis using perdeuterated protein
IUCrJ, 1, 2014
|
|
4PVN
 
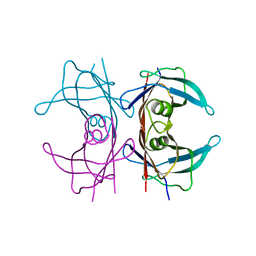 | | Neutron structure of human transthyretin (TTR) at room temperature to 2.3A resolution (monochromatic) | | Descriptor: | Transthyretin | | Authors: | Fisher, S.J, Blakeley, M.P, Haupt, M, Mason, S.A, Cooper, J.B, Mitchell, E.P, Forsyth, V.T. | | Deposit date: | 2014-03-18 | | Release date: | 2014-11-12 | | Last modified: | 2024-03-20 | | Method: | NEUTRON DIFFRACTION (2.3 Å), X-RAY DIFFRACTION | | Cite: | Binding site asymmetry in human transthyretin: insights from a joint neutron and X-ray crystallographic analysis using perdeuterated protein
IUCrJ, 1, 2014
|
|
4UFT
 
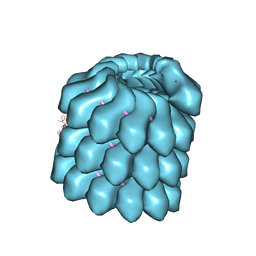 | | Structure of the helical Measles virus nucleocapsid | | Descriptor: | 5'-R(*CP*CP*CP*CP*CP*CP)-3', NUCLEOPROTEIN | | Authors: | Gutsche, I, Desfosses, A, Effantin, G, Ling, W.L, Haupt, M, Ruigrok, R.W.H, Sachse, C, Schoehn, G. | | Deposit date: | 2015-03-19 | | Release date: | 2015-04-29 | | Last modified: | 2019-04-24 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Near-Atomic Cryo-Em Structure of the Helical Measles Virus Nucleocapsid.
Science, 348, 2015
|
|
2J04
 
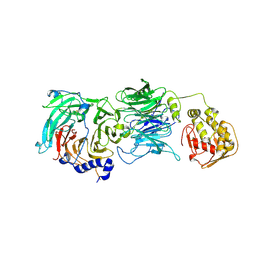 | | The tau60-tau91 subcomplex of yeast transcription factor IIIC | | Descriptor: | HYPOTHETICAL PROTEIN YPL007C, YDR362CP | | Authors: | Mylona, A, Fernandez-Tornero, C, Legrand, P, Muller, C.W. | | Deposit date: | 2006-07-31 | | Release date: | 2006-10-23 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the Tau60/Deltatau91 Subcomplex of Yeast Transcription Factor Iiic: Insights Into Preinitiation Complex Assembly
Mol.Cell, 24, 2006
|
|
