5XN8
 
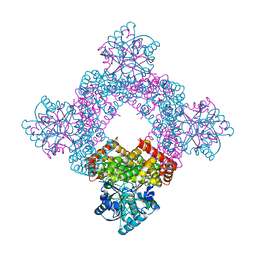 | | Structure of glycerol dehydrogenase crystallised as a contaminant | | Descriptor: | GLYCEROL, Glycerol Dehydrogenase, ZINC ION | | Authors: | Hatti, K, Mathiharan, Y.K, Srinivasan, N, Murthy, M.R.N. | | Deposit date: | 2017-05-19 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Seeing but not believing: the structure of glycerol dehydrogenase initially assumed to be the structure of a survival protein from Salmonella typhimurium
Acta Crystallogr.,Sect.D, 73, 2017
|
|
5H3L
 
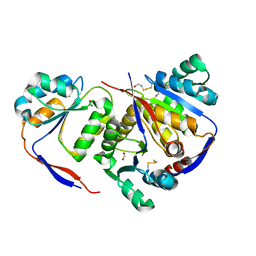 | | Structure of methylglyoxal synthase crystallised as a contaminant | | Descriptor: | FORMIC ACID, Methylglyoxal synthase | | Authors: | Hatti, K, Dadireddy, V, Srinivasan, N, Ramakumar, S, Murthy, M.R.N. | | Deposit date: | 2016-10-25 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure determination of contaminant proteins using the MarathonMR procedure.
J. Struct. Biol., 197, 2017
|
|
5J1D
 
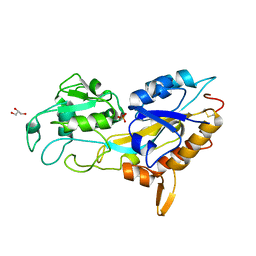 | | X-ray crystal structure of Phosphate binding protein (PBP) from Stenotrophomonas maltophilia | | Descriptor: | GLYCEROL, PHOSPHATE ION, Phosphate binding protein | | Authors: | Hatti, K, Gulati, A, Narayanswamy, S, Murthy, M.R.N. | | Deposit date: | 2016-03-29 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of crystal structures of proteins of unknown identity using a marathon molecular replacement procedure: structure of Stenotrophomonas maltophilia phosphate-binding protein.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5H4G
 
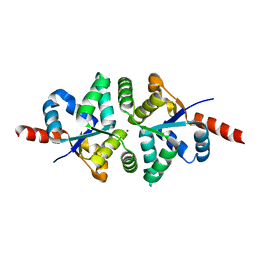 | | Structure of PIN-domain protein (VapC4 toxin) from Pyrococcus horikoshii determined at 1.77 A resolution | | Descriptor: | Ribonuclease VapC4, ZINC ION | | Authors: | Biswas, A, Hatti, K, Srinivasan, N, Murthy, M.R.N, Sekar, K. | | Deposit date: | 2016-10-31 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure determination of contaminant proteins using the MarathonMR procedure
J. Struct. Biol., 197, 2017
|
|
5H4H
 
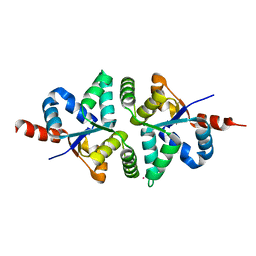 | | Structure of PIN-domain protein (VapC4 toxin) from Pyrococcus horikoshii determined at 2.2 A resolution | | Descriptor: | CADMIUM ION, Ribonuclease VapC4 | | Authors: | Biswas, A, Hatti, K, Srinivasan, N, Murthy, M.R.N, Sekar, K. | | Deposit date: | 2016-10-31 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure determination of contaminant proteins using the MarathonMR procedure
J. Struct. Biol., 197, 2017
|
|
5H4F
 
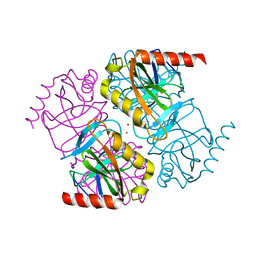 | | Structure of inorganic pyrophosphatase crystallised as a contaminant | | Descriptor: | ZINC ION, inorganic pyrophosphatase | | Authors: | Chaudhary, S, Hatti, K, Srinivasan, N, Murthy, M.R.N, Sekar, K. | | Deposit date: | 2016-10-31 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure determination of contaminant proteins using the MarathonMR procedure.
J. Struct. Biol., 197, 2017
|
|
