1IYY
 
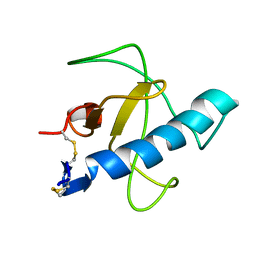 | | NMR STRUCTURE OF Gln25-RIBONUCLEASE T1, 24 STRUCTURES | | Descriptor: | RIBONUCLEASE T1 | | Authors: | Hatano, K, Kojima, M, Suzuki, E, Tanokura, M, Takahashi, K. | | Deposit date: | 2002-09-12 | | Release date: | 2003-10-07 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Determination of the NMR structure of Gln25-ribonuclease T1.
Biol. Chem., 384, 2003
|
|
1X25
 
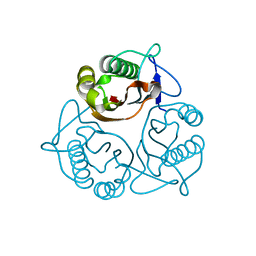 | | Crystal Structure of a Member of YjgF Family from Sulfolobus Tokodaii (ST0811) | | Descriptor: | Hypothetical UPF0076 protein ST0811 | | Authors: | Miyakawa, T, Lee, W.C, Hatano, K, Kato, Y, Sawano, Y, Miyazono, K, Nagata, K, Tanokura, M. | | Deposit date: | 2005-04-20 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the YjgF/YER057c/UK114 family protein from the hyperthermophilic archaeon Sulfolobus tokodaii strain 7
Proteins, 62, 2006
|
|
4XRE
 
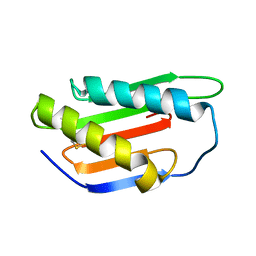 | | Crystal structure of Gnk2 complexed with mannose | | Descriptor: | Antifungal protein ginkbilobin-2, alpha-D-mannopyranose | | Authors: | Miyakawa, T, Hatano, K, Miyauchi, Y, Suwa, Y, Sawano, Y, Tanokura, M. | | Deposit date: | 2015-01-21 | | Release date: | 2015-02-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | A secreted protein with plant-specific cysteine-rich motif functions as a mannose-binding lectin that exhibits antifungal activity.
Plant Physiol., 166, 2014
|
|
5X1B
 
 | | CO bound cytochrome c oxidase at 20 nsec after pump laser irradiation to release CO from O2 reduction center | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
5X19
 
 | | CO bound cytochrome c oxidase at 100 micro sec after pump laser irradiation to release CO from O2 reduction center | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
5X1F
 
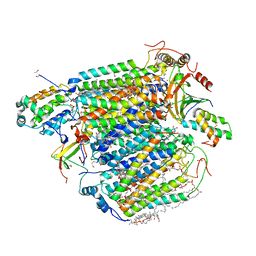 | | CO bound cytochrome c oxidase without pump laser irradiation at 278K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
2EJX
 
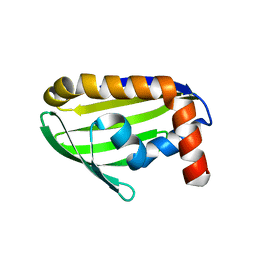 | | Crystal structure of the hypothetical protein STK_08120 from Sulfolobus tokodaii | | Descriptor: | STK_08120 | | Authors: | Miyakawa, T, Miyazono, K, Sawano, Y, Hatano, K, Nagata, K, Tanokura, M. | | Deposit date: | 2007-03-21 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A thermoacidophile-specific protein family, DUF3211, functions as a fatty acid carrier with novel binding mode
J.Bacteriol., 195, 2013
|
|
3W9K
 
 | | Crystal structure of thermoacidophile-specific protein STK_08120 complexed with myristic acid | | Descriptor: | FATTY ACID-BINDING PROTEIN, MYRISTIC ACID | | Authors: | Miyakawa, T, Sawano, Y, Miyazono, K, Miyauchi, Y, Hatano, K, Tanokura, M. | | Deposit date: | 2013-04-05 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A thermoacidophile-specific protein family, DUF3211, functions as a fatty acid carrier with novel binding mode.
J.Bacteriol., 195, 2013
|
|
3A2E
 
 | | Crystal structure of ginkbilobin-2, the novel antifungal protein from Ginkgo biloba seeds | | Descriptor: | Ginkbilobin-2 | | Authors: | Miyakawa, T, Miyazono, K, Sawano, Y, Hatano, K, Tanokura, M. | | Deposit date: | 2009-05-13 | | Release date: | 2009-06-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of ginkbilobin-2 with homology to the extracellular domain of plant cysteine-rich receptor-like kinases
Proteins, 77, 2009
|
|
5ZCP
 
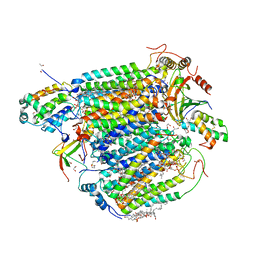 | | azide-bound cytochrome c oxidase structure determined using the crystals exposed to 20 mM azide solution for 2 days | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Hatano, K, Tadehara, H, Tsukihara, T. | | Deposit date: | 2018-02-19 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity.
J. Biol. Chem., 293, 2018
|
|
5Z86
 
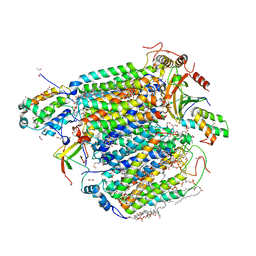 | | azide-bound cytochrome c oxidase structure determined using the crystals exposed to 20 mM azide solution for 3 days | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Hatano, K, Tadehara, H, Tsukihara, T. | | Deposit date: | 2018-01-31 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity.
J. Biol. Chem., 293, 2018
|
|
5ZCQ
 
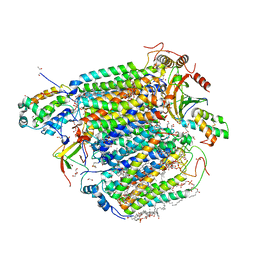 | | Azide-bound cytochrome c oxidase structure determined using the crystals exposed to 10 mM azide solution for 2 days | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Hatano, K, Tadehara, H, Tsukihara, T. | | Deposit date: | 2018-02-19 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity.
J. Biol. Chem., 293, 2018
|
|
5ZCO
 
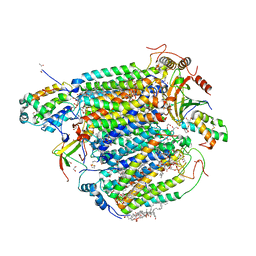 | | azide-bound cytochrome c oxidase structure determined using the crystals exposed to 2 mM azide solution for 2 days | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Hatano, K, Tadehara, H, Tsukihara, T. | | Deposit date: | 2018-02-19 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity.
J. Biol. Chem., 293, 2018
|
|
5Z84
 
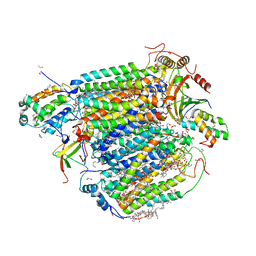 | | The structure of azide-bound cytochrome c oxidase determined using the crystals exposed to 20 mM azide solution for 4 days | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Hatano, K, Tadehara, H, Tsukihara, T. | | Deposit date: | 2018-01-31 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity.
J. Biol. Chem., 293, 2018
|
|
5Z85
 
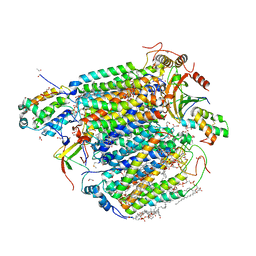 | | The structure of azide-bound cytochrome c oxidase determined using the another batch crystals exposed to 20 mM azide solution for 2 days | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Hatano, K, Tadehara, H, Tsukihara, T. | | Deposit date: | 2018-01-31 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity.
J. Biol. Chem., 293, 2018
|
|
1BI6
 
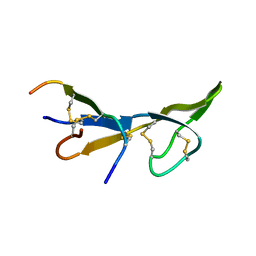 | | NMR STRUCTURE OF BROMELAIN INHIBITOR VI FROM PINEAPPLE STEM | | Descriptor: | BROMELAIN INHIBITOR VI | | Authors: | Hatano, K.-I. | | Deposit date: | 1995-12-07 | | Release date: | 1996-04-03 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of bromelain inhibitor IV from pineapple stem: structural similarity with Bowman-Birk trypsin/chymotrypsin inhibitor from soybean.
Biochemistry, 35, 1996
|
|
2BI6
 
 | | NMR STUDY OF BROMELAIN INHIBITOR VI FROM PINEAPPLE STEM | | Descriptor: | BROMELAIN INHIBITOR VI | | Authors: | Hatano, K.-I. | | Deposit date: | 1995-12-07 | | Release date: | 1996-04-03 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of bromelain inhibitor IV from pineapple stem: structural similarity with Bowman-Birk trypsin/chymotrypsin inhibitor from soybean.
Biochemistry, 35, 1996
|
|
8GVM
 
 | | The structure of azide-bound cytochrome C oxidase determined using the crystals exposed to 20 mm azide solution for 4 days | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimada, A. | | Deposit date: | 2022-09-15 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | X-ray structural analyses of azide-bound cytochromecoxidases reveal that the H-pathway is critically important for the proton-pumping activity
J. Biol. Chem., 293, 2018
|
|
4TWL
 
 | | Crystal structure of dioscorin complexed with ascorbate | | Descriptor: | ASCORBIC ACID, Dioscorin 5, SULFATE ION | | Authors: | Xue, Y.L, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2014-07-01 | | Release date: | 2015-04-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Yam Tuber Storage Protein Reduces Plant Oxidants Using the Coupled Reactions as Carbonic Anhydrase and Dehydroascorbate Reductase
Mol Plant, 8, 2015
|
|
4TWM
 
 | | Crystal structure of dioscorin from Dioscorea japonica | | Descriptor: | Dioscorin 5, SULFATE ION | | Authors: | Xue, Y.L, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2014-07-01 | | Release date: | 2015-04-01 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Yam Tuber Storage Protein Reduces Plant Oxidants Using the Coupled Reactions as Carbonic Anhydrase and Dehydroascorbate Reductase
Mol Plant, 8, 2015
|
|
