2OQ1
 
 | | Tandem SH2 domains of ZAP-70 with 19-mer zeta1 peptide | | Descriptor: | LEAD (II) ION, T-cell surface glycoprotein CD3 zeta chain, Tyrosine-protein kinase ZAP-70 | | Authors: | Hatada, M.H, Laird, E.R, Green, J, Morgenstern, J, Ram, M.K. | | Deposit date: | 2007-01-30 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for the interaction of ZAP-70 with the T-cell receptor
Nature, 377, 1995
|
|
1HXB
 
 | | HIV-1 proteinase complexed with RO 31-8959 | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, HIV-1 PROTEASE | | Authors: | Graves, B.J, Hatada, M.H, Crowther, R.L. | | Deposit date: | 1996-09-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel binding mode of highly potent HIV-proteinase inhibitors incorporating the (R)-hydroxyethylamine isostere.
J.Med.Chem., 34, 1991
|
|
2ILA
 
 | |
1BL4
 
 | | FKBP MUTANT F36V COMPLEXED WITH REMODELED SYNTHETIC LIGAND | | Descriptor: | PROTEIN (FK506 BINDING PROTEIN), {3-[3-(3,4-DIMETHOXY-PHENYL)-1-(1-{1-[2-(3,4,5-TRIMETHOXY-PHENYL)-BUTYRYL]-PIPERIDIN-2YL}-VINYLOXY)-PROPYL]-PHENOXY}-ACETIC ACID | | Authors: | Hatada, M.H, Clackson, T, Yang, W, Rozamus, L.W, Amara, J, Rollins, C.T, Stevenson, L.F, Magari, S.R, Wood, S.A, Courage, N.L, Lu, X, Cerasoli Junior, F, Gilman, M, Holt, D. | | Deposit date: | 1998-07-23 | | Release date: | 1998-09-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1KGA
 
 | |
1CSZ
 
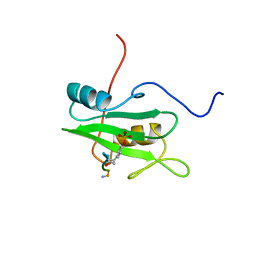 | | SYK TYROSINE KINASE C-TERMINAL SH2 DOMAIN COMPLEXED WITH A PHOSPHOPEPTIDEFROM THE GAMMA CHAIN OF THE HIGH AFFINITY IMMUNOGLOBIN G RECEPTOR, NMR | | Descriptor: | ACETYL-THR-PTR-GLU-THR-LEU-NH2, SYK PROTEIN TYROSINE KINASE | | Authors: | Narula, S.S, Yuan, R.W, Adams, S.E, Green, O.M, Green, J, Phillips, T.B, Zydowsky, L.D, Botfield, M.C, Hatada, M.H, Laird, E.R, Zoller, M.J, Karas, J.L, Dalgarno, D.C. | | Deposit date: | 1995-10-03 | | Release date: | 1996-11-08 | | Last modified: | 2012-02-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal SH2 domain of the human tyrosine kinase Syk complexed with a phosphotyrosine pentapeptide.
Structure, 3, 1995
|
|
1CSY
 
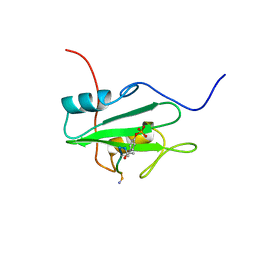 | | SYK TYROSINE KINASE C-TERMINAL SH2 DOMAIN COMPLEXED WITH A PHOSPHOPEPTIDEFROM THE GAMMA CHAIN OF THE HIGH AFFINITY IMMUNOGLOBIN G RECEPTOR, NMR | | Descriptor: | ACETYL-THR-PTR-GLU-THR-LEU-NH2, SYK PROTEIN TYROSINE KINASE | | Authors: | Narula, S.S, Yuan, R.W, Adams, S.E, Green, O.M, Green, J, Phillips, T.B, Zydowsky, L.D, Botfield, M.C, Hatada, M.H, Laird, E.R, Zoller, M.J, Karas, J.L, Dalgarno, D.C. | | Deposit date: | 1995-10-03 | | Release date: | 1996-11-08 | | Last modified: | 2012-02-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal SH2 domain of the human tyrosine kinase Syk complexed with a phosphotyrosine pentapeptide.
Structure, 3, 1995
|
|
1C6Z
 
 | | ALTERNATE BINDING SITE FOR THE P1-P3 GROUP OF A CLASS OF POTENT HIV-1 PROTEASE INHIBITORS AS A RESULT OF CONCERTED STRUCTURAL CHANGE IN 80'S LOOP. | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, PROTEIN (PROTEASE) | | Authors: | Munshi, S. | | Deposit date: | 1999-12-28 | | Release date: | 2000-12-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An alternate binding site for the P1-P3 group of a class of potent HIV-1 protease inhibitors as a result of concerted structural change in the 80s loop of the protease.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1MXS
 
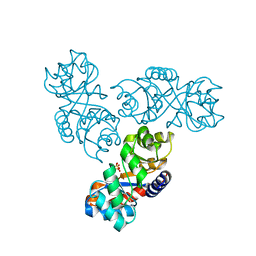 | | Crystal structure of 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolase from Pseudomonas putida. | | Descriptor: | KDPG Aldolase, SULFATE ION | | Authors: | Watanabe, L, Bell, B.J, Lebioda, L, Rios-Steiner, J.L, Tulinsky, A, Arni, R.K. | | Deposit date: | 2002-10-03 | | Release date: | 2003-09-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolase from Pseudomonas putida.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
