4EH1
 
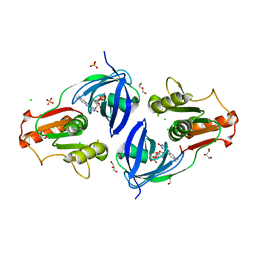 | | Crystal Structure of the Flavohem-like-FAD/NAD Binding Domain of Nitric Oxide Dioxygenase from Vibrio cholerae O1 biovar El Tor | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Flavohemoprotein, ... | | Authors: | Kim, Y, Gu, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-02 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Flavohem-like-FAD/NAD Binding Domain of Nitric Oxide Dioxygenase from Vibrio cholerae O1 biovar El Tor
To be Published
|
|
3DZC
 
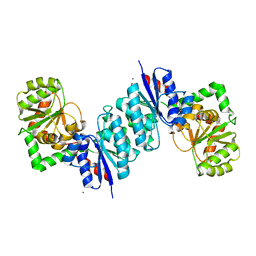 | | 2.35 Angstrom resolution structure of WecB (VC0917), a UDP-N-acetylglucosamine 2-epimerase from Vibrio cholerae. | | Descriptor: | CALCIUM ION, CHLORIDE ION, UDP-N-acetylglucosamine 2-epimerase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Kwon, K, Hasseman, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-07-29 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom resolution structure of WecB (VC0917), a UDP-N-acetylglucosamine 2-epimerase from Vibrio cholerae.
TO BE PUBLISHED
|
|
3ECT
 
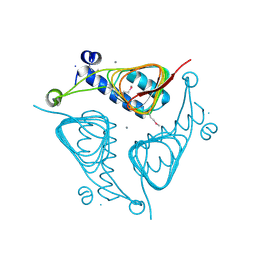 | | Crystal Structure of the Hexapeptide-Repeat Containing-Acetyltransferase VCA0836 from Vibrio cholerae | | Descriptor: | CALCIUM ION, Hexapeptide-repeat containing-acetyltransferase | | Authors: | Kim, Y, Maltseva, N, Kwon, K, Papazisi, L, Hasseman, J, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-02 | | Release date: | 2008-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of the Hexapeptide-Repeat Containing-Acetyltransferase VCA0836 from Vibrio cholerae
To be Published
|
|
3L07
 
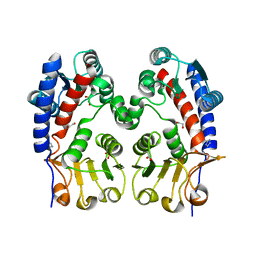 | | Methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase, putative bifunctional protein folD from Francisella tularensis. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Bifunctional protein folD, ... | | Authors: | Osipiuk, J, Maltseva, N, Mulligan, R, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-09 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | X-ray crystal structure of methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase, putative bifunctional protein folD from Francisella tularensis.
To be Published
|
|
3LJK
 
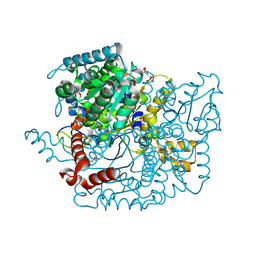 | | Glucose-6-phosphate isomerase from Francisella tularensis. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Osipiuk, J, Maltseva, N, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-26 | | Release date: | 2010-03-16 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | X-ray crystal structure of glucose-6-phosphate isomerase from Francisella tularensis.
To be Published
|
|
3M5P
 
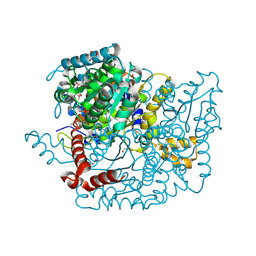 | | Glucose-6-phosphate isomerase from Francisella tularensis complexed with fructose-6-phosphate. | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, GLYCEROL, Glucose-6-phosphate isomerase, ... | | Authors: | Osipiuk, J, Maltseva, N, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-12 | | Release date: | 2010-03-23 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray crystal structure of glucose-6-phosphate isomerase from Francisella tularensis complexed with fructose-6-phosphate.
To be Published
|
|
3Q88
 
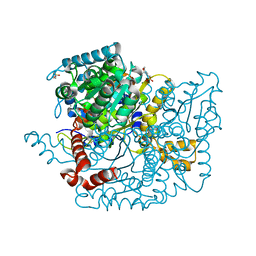 | | Glucose-6-phosphate isomerase from Francisella tularensis complexed with ribose 1,5-bisphosphate. | | Descriptor: | 1,5-di-O-phosphono-alpha-D-ribofuranose, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Osipiuk, J, Maltseva, N, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Glucose-6-phosphate isomerase from Francisella tularensis.
To be Published
|
|
3Q7I
 
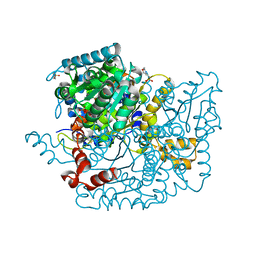 | | Glucose-6-phosphate isomerase from Francisella tularensis complexed with 6-phosphogluconic acid. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-PHOSPHOGLUCONIC ACID, CALCIUM ION, ... | | Authors: | Osipiuk, J, Maltseva, N, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-04 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Glucose-6-phosphate isomerase from Francisella tularensis.
To be Published
|
|
3RHI
 
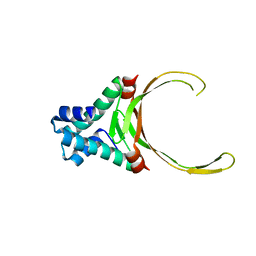 | | DNA-binding protein HU from Bacillus anthracis | | Descriptor: | DNA-binding protein HU | | Authors: | Osipiuk, J, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-11 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | DNA-binding protein HU from Bacillus anthracis.
To be Published
|
|
3L44
 
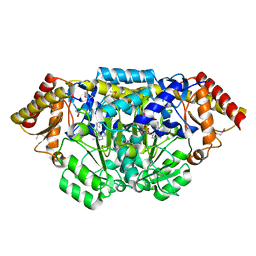 | | Crystal structure of Bacillus anthracis HemL-1, glutamate semialdehyde aminotransferase | | Descriptor: | Glutamate-1-semialdehyde 2,1-aminomutase 1 | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Hasseman, J, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Bacillus anthracis HemL-1, glutamate semialdehyde aminotransferase
TO BE PUBLISHED
|
|
3LER
 
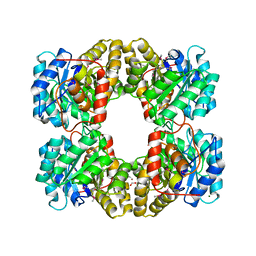 | | Crystal Structure of Dihydrodipicolinate Synthase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Zhou, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-15 | | Release date: | 2010-01-26 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure of Dihydrodipicolinate Synthase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
3LQN
 
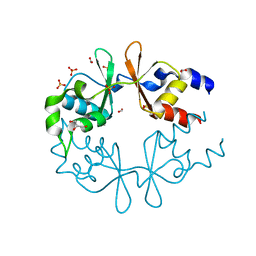 | | Crystal Structure of CBS Domain-containing Protein of Unknown Function from Bacillus anthracis str. Ames Ancestor | | Descriptor: | CBS domain protein, FORMIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Mulligan, R, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-09 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of CBS Domain-containing Protein of Unknown Function from Bacillus anthracis str. Ames Ancestor
To be Published
|
|
3LAC
 
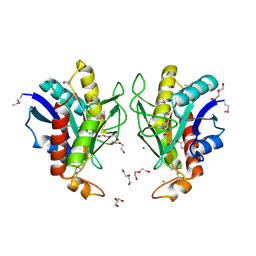 | | Crystal structure of Bacillus anthracis pyrrolidone-carboxylate peptidase, pcP | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Pyrrolidone-carboxylate peptidase | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Hasseman, J, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-06 | | Release date: | 2010-01-19 | | Last modified: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Bacillus anthracis pyrrolidone-carboxylate peptidase, pcP
To be Published
|
|
3LU2
 
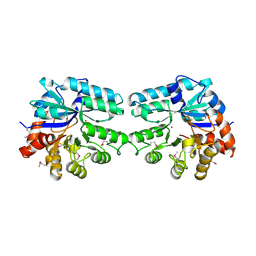 | | Structure of lmo2462, a Listeria monocytogenes amidohydrolase family putative dipeptidase | | Descriptor: | Lmo2462 protein, ZINC ION | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Hasseman, J, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-16 | | Release date: | 2010-03-09 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of lmo2462, a Listeria monocytogenes amidohydrolase family putative dipeptidase
To be Published
|
|
3LYL
 
 | | Structure of 3-oxoacyl-acylcarrier protein reductase, FabG from Francisella tularensis | | Descriptor: | 3-oxoacyl-(Acyl-carrier-protein) reductase | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Hasseman, J, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-27 | | Release date: | 2010-03-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of 3-oxoacyl-acylcarrier protein reductase, FabG from Francisella tularensis
TO BE PUBLISHED
|
|
3M5W
 
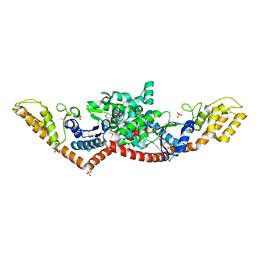 | | Crystal Structure of Tryptophanyl-tRNA Synthetase from Campylobacter jejuni | | Descriptor: | GLYCEROL, SULFATE ION, Tryptophanyl-tRNA synthetase | | Authors: | Kim, Y, Zhou, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-14 | | Release date: | 2010-03-31 | | Last modified: | 2014-10-01 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal Structure of Tryptophanyl-tRNA Synthetase from Campylobacter jejuni
To be Published, 2010
|
|
3MSU
 
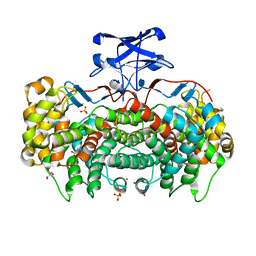 | | Crystal Structure of Citrate Synthase from Francisella tularensis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Maltseva, N, Kim, Y, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-04-29 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.843 Å) | | Cite: | Crystal Structure of Citrate Synthase from Francisella tularensis
To be Published
|
|
3M84
 
 | | Crystal Structure of Phosphoribosylaminoimidazole Synthetase from Francisella tularensis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Maltseva, N, Kim, Y, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-17 | | Release date: | 2010-07-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal Structure of Phosphoribosylaminoimidazole Synthetase from
Francisella tularensis
To be Published
|
|
3N0L
 
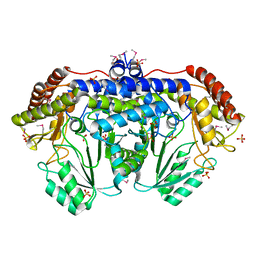 | | Crystal structure of serine hydroxymethyltransferase from Campylobacter jejuni | | Descriptor: | SULFATE ION, Serine hydroxymethyltransferase | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Hasseman, J, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-14 | | Release date: | 2010-05-26 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of serine hydroxymethyltransferase from Campylobacter jejuni
TO BE PUBLISHED
|
|
3O6V
 
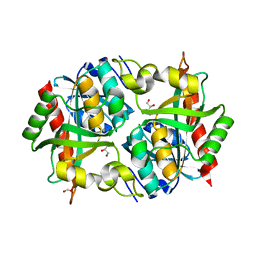 | | Crystal structure of Uridine Phosphorylase from Vibrio cholerae O1 biovar El Tor | | Descriptor: | FORMIC ACID, GLYCEROL, Uridine phosphorylase | | Authors: | Maltseva, N, Kim, Y, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-29 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Crystal structure of Uridine Phosphorylase from Vibrio cholerae O1 biovar El Tor
To be Published
|
|
3NTX
 
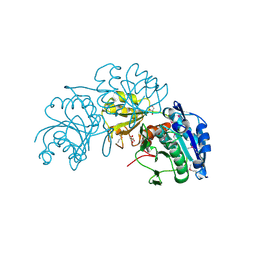 | | Crystal Structure of L-asparaginase I from Yersinia pestis | | Descriptor: | Cytoplasmic L-asparaginase I, GLYCEROL | | Authors: | Kim, Y, Gu, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-05 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of L-asparaginase I from Yersinia pestis
To be Published, 2010
|
|
3NZ2
 
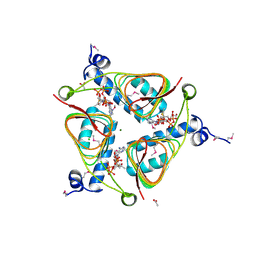 | | Crystal Structure of Hexapeptide-Repeat containing-Acetyltransferase VCA0836 Complexed with Acetyl Co Enzyme A from Vibrio cholerae O1 biovar eltor | | Descriptor: | 1,4-BUTANEDIOL, ACETIC ACID, ACETYL COENZYME *A, ... | | Authors: | Kim, Y, Maltseva, N, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-15 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Hexapeptide-Repeat containing-Acetyltransferase VCA0836 Complexed with Acetyl Co Enzyme A from Vibrio cholerae O1 biovar eltor
To be Published
|
|
3P09
 
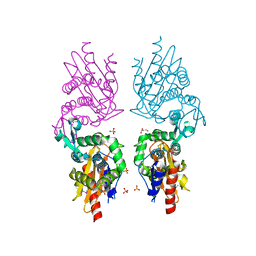 | | Crystal Structure of Beta-Lactamase from Francisella tularensis | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Kim, Y, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-27 | | Release date: | 2010-10-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal Structure of Beta-Lactamase from Francisella tularensis
To be Published
|
|
3PNS
 
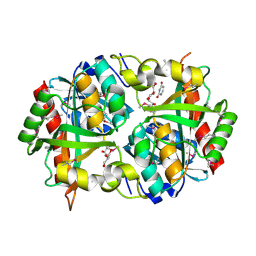 | | Crystal Structure of Uridine Phosphorylase Complexed with Uracil from Vibrio cholerae O1 biovar El Tor | | Descriptor: | ACETIC ACID, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-11-19 | | Release date: | 2010-12-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Crystal Structure of Uridine Phosphorylase Complexed with Uracil from Vibrio cholerae O1 biovar El Tor
To be Published
|
|
3Q7H
 
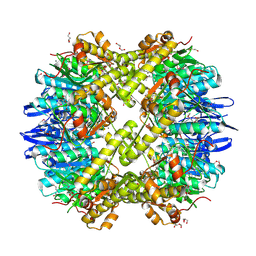 | | Structure of the ClpP subunit of the ATP-dependent Clp Protease from Coxiella burnetii | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CALCIUM ION, DI(HYDROXYETHYL)ETHER | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Hasseman, J, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the ClpP subunit of the ATP-dependent Clp Protease from Coxiella burnetii
To be Published
|
|
