1ZNM
 
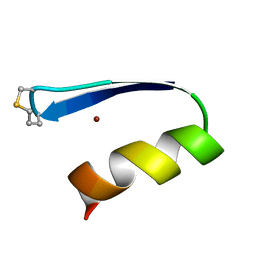 | | A zinc finger with an artificial beta-turn, original sequence taken from the third zinc finger domain of the human transcriptional repressor protein YY1 (YING and YANG 1, a delta transcription factor), nmr, 34 structures | | Descriptor: | YY1, ZINC ION | | Authors: | Viles, J.H, Patel, S.U, Mitchell, J.B.O, Moody, C.M, Justice, D.E, Uppenbrink, J, Doyle, P.M, Harris, C.J, Sadler, P.J, Thornton, J.M. | | Deposit date: | 1997-11-20 | | Release date: | 1998-04-01 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Design, synthesis and structure of a zinc finger with an artificial beta-turn.
J.Mol.Biol., 279, 1998
|
|
5ER1
 
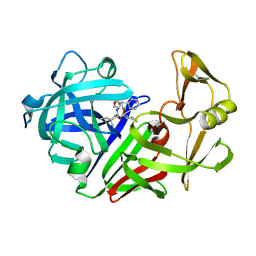 | |
6T28
 
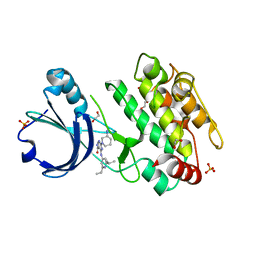 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 19 (CS640) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[2,6-di(propan-2-yl)pyridin-4-yl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Sorrell, F, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-08 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Highly Selective Inhibitors of Calmodulin-Dependent Kinases That Restore Insulin Sensitivity in the Diet-Induced Obesityin VivoMouse Model.
J.Med.Chem., 63, 2020
|
|
6T29
 
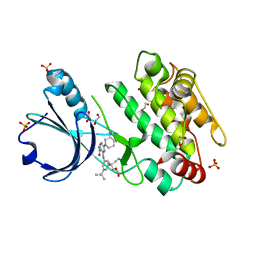 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 18 (CS587) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[3,5-bis(2-cyanopropan-2-yl)phenyl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Sorrell, F, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-08 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.484 Å) | | Cite: | Discovery of Highly Selective Inhibitors of Calmodulin-Dependent Kinases That Restore Insulin Sensitivity in the Diet-Induced Obesityin VivoMouse Model.
J.Med.Chem., 63, 2020
|
|
7VG3
 
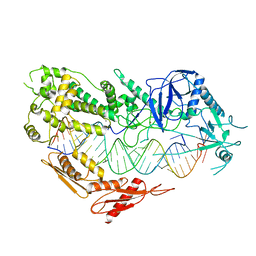 | | Cryo-EM structure of Arabidopsis DCL3 in complex with a 30-bp RNA | | Descriptor: | CALCIUM ION, Dicer-like 3, TAS1a RNA forward strand (5'-phosphorylated), ... | | Authors: | Wang, Q, Du, J. | | Deposit date: | 2021-09-14 | | Release date: | 2021-10-27 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Mechanism of siRNA production by a plant Dicer-RNA complex in dicing-competent conformation.
Science, 374, 2021
|
|
7VG2
 
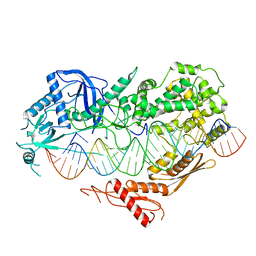 | | Cryo-EM structure of Arabidopsis DCL3 in complex with a 40-bp RNA | | Descriptor: | CALCIUM ION, Dicer-like 3, TAS1a forward strand (5'-phosphorylation), ... | | Authors: | Wang, Q, Du, J. | | Deposit date: | 2021-09-14 | | Release date: | 2021-10-27 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of siRNA production by a plant Dicer-RNA complex in dicing-competent conformation.
Science, 374, 2021
|
|
6A5M
 
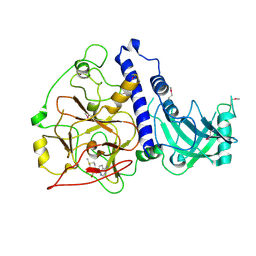 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with SAM, form 2 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6, S-ADENOSYLMETHIONINE, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6A5N
 
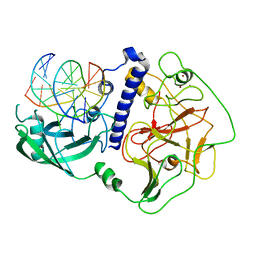 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with methylated DNA | | Descriptor: | DNA (5'-D(*CP*AP*CP*TP*GP*CP*TP*GP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*GP*AP*GP*TP*AP*CP*TP*(5CM)P*AP*GP*CP*AP*GP*T)-3'), Histone-lysine N-methyltransferase, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6A5K
 
 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with SAM, form 1 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6, S-ADENOSYLMETHIONINE, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
