2BYN
 
 | | Crystal structure of apo AChBP from Aplysia californica | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PENTAETHYLENE GLYCOL, SOLUBLE ACETYLCHOLINE RECEPTOR, ... | | Authors: | Hansen, S.B, Sulzenbacher, G, Huxford, T, Marchot, P, Taylor, P, Bourne, Y. | | Deposit date: | 2005-08-03 | | Release date: | 2005-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structures of Aplysia Achbp Complexes with Nicotinic Agonists and Antagonists Reveal Distinctive Binding Interfaces and Conformations.
Embo J., 24, 2005
|
|
2BYS
 
 | | CRYSTAL STRUCTURE OF ACHBP FROM APLYSIA CALIFORNICA IN complex with lobeline | | Descriptor: | ACETYLCHOLINE-BINDING PROTEIN, LOBELINE | | Authors: | Hansen, S.B, Sulzenbacher, G, Huxford, T, Marchot, P, Taylor, P, Bourne, Y. | | Deposit date: | 2005-08-04 | | Release date: | 2005-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of Aplysia Achbp Complexes with Nicotinic Agonists and Antagonists Reveal Distinctive Binding Interfaces and Conformations.
Embo J., 24, 2005
|
|
2BYP
 
 | | Crystal structure of Aplysia californica AChBP in complex with alpha- conotoxin ImI | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-CONOTOXIN IMI, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Hansen, S.B, Sulzenbacher, G, Huxford, T, Marchot, P, Taylor, P, Bourne, Y. | | Deposit date: | 2005-08-03 | | Release date: | 2005-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structures of Aplysia Achbp Complexes with Nicotinic Agonists and Antagonists Reveal Distinctive Binding Interfaces and Conformations.
Embo J., 24, 2005
|
|
2PGZ
 
 | | Crystal structure of Cocaine bound to an ACh-Binding Protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COCAINE, Soluble acetylcholine receptor, ... | | Authors: | Hansen, S.B, Taylor, P. | | Deposit date: | 2007-04-10 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Galanthamine and non-competitive inhibitor binding to ACh-binding protein: evidence for a binding site on non-alpha-subunit interfaces of heteromeric neuronal nicotinic receptors.
J.Mol.Biol., 369, 2007
|
|
2BYR
 
 | | CRYSTAL STRUCTURE OF ACHBP FROM APLYSIA CALIFORNICA in complex with methyllycaconitine | | Descriptor: | ACETYLCHOLINE-BINDING PROTEIN, METHYLLYCACONITINE | | Authors: | Hansen, S.B, Sulzenbacher, G, Huxford, T, Marchot, P, Taylor, P, Bourne, Y. | | Deposit date: | 2005-08-03 | | Release date: | 2005-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of Aplysia Achbp Complexes with Nicotinic Agonists and Antagonists Reveal Distinctive Binding Interfaces and Conformations.
Embo J., 24, 2005
|
|
2BYQ
 
 | | Crystal structure of Aplysia californica AChBP in complex with epibatidine | | Descriptor: | EPIBATIDINE, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Hansen, S.B, Sulzenbacher, G, Huxford, T, Marchot, P, Taylor, P, Bourne, Y. | | Deposit date: | 2005-08-03 | | Release date: | 2005-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of Aplysia Achbp Complexes with Nicotinic Agonists and Antagonists Reveal Distinctive Binding Interfaces and Conformations.
Embo J., 24, 2005
|
|
2PH9
 
 | | Galanthamine bound to an ACh-binding Protein | | Descriptor: | (-)-GALANTHAMINE, Soluble acetylcholine receptor, TETRAETHYLENE GLYCOL | | Authors: | Hansen, S.B, Taylor, P. | | Deposit date: | 2007-04-10 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Galanthamine and non-competitive inhibitor binding to ACh-binding protein: evidence for a binding site on non-alpha-subunit interfaces of heteromeric neuronal nicotinic receptors.
J.Mol.Biol., 369, 2007
|
|
3SPI
 
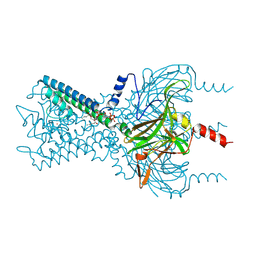 | | Inward rectifier potassium channel Kir2.2 in complex with PIP2 | | Descriptor: | Inward-rectifier K+ channel Kir2.2, POTASSIUM ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Hansen, S.B, Tao, X, MacKinnon, R. | | Deposit date: | 2011-07-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.307 Å) | | Cite: | Structural basis of PIP(2) activation of the classical inward rectifier K(+) channel Kir2.2.
Nature, 477, 2011
|
|
3SPH
 
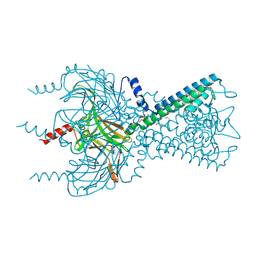 | | Inward rectifier potassium channel Kir2.2 I223L mutant in complex with PIP2 | | Descriptor: | Inward-rectifier K+ channel Kir2.2, POTASSIUM ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Hansen, S.B, Tao, X, MacKinnon, R. | | Deposit date: | 2011-07-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structural basis of PIP(2) activation of the classical inward rectifier K(+) channel Kir2.2.
Nature, 477, 2011
|
|
3SPC
 
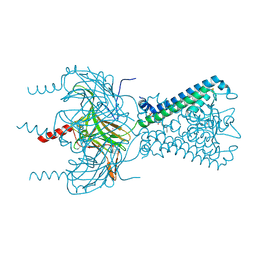 | | Inward rectifier potassium channel Kir2.2 in complex with dioctanoylglycerol pyrophosphate (DGPP) | | Descriptor: | (2R)-3-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}propane-1,2-diyl dioctanoate, Inward-rectifier K+ channel Kir2.2, POTASSIUM ION | | Authors: | Hansen, S.B, Tao, X, MacKinnon, R. | | Deposit date: | 2011-07-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.454 Å) | | Cite: | Structural basis of PIP(2) activation of the classical inward rectifier K(+) channel Kir2.2.
Nature, 477, 2011
|
|
3SPG
 
 | | Inward rectifier potassium channel Kir2.2 R186A mutant in complex with PIP2 | | Descriptor: | Inward-rectifier K+ channel Kir2.2, POTASSIUM ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Hansen, S.B, Tao, X, MacKinnon, R. | | Deposit date: | 2011-07-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.613 Å) | | Cite: | Structural basis of PIP(2) activation of the classical inward rectifier K(+) channel Kir2.2.
Nature, 477, 2011
|
|
3SPJ
 
 | |
5NLW
 
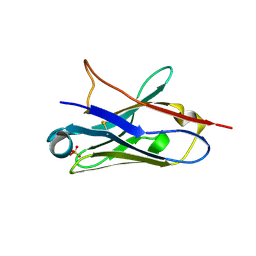 | | Structure of Nb36 crystal form 2 | | Descriptor: | SULFATE ION, nanobody Nb36 | | Authors: | Hansen, S.B, Andersen, K.R, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-31 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Introducing site-specific cysteines into nanobodies for mercury labelling allows de novo phasing of their crystal structures.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5NML
 
 | | Nb36 Ser85Cys with Hg bound | | Descriptor: | 1,2-ETHANEDIOL, MERCURY (II) ION, Nanobody Nb36 Ser85Cys | | Authors: | Hansen, S.B, Andersen, K.R, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2017-04-06 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Introducing site-specific cysteines into nanobodies for mercury labelling allows de novo phasing of their crystal structures.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5NM0
 
 | | Nb36 Ser85Cys with Hg, crystal form 1 | | Descriptor: | MERCURY (II) ION, Nb36 | | Authors: | Hansen, S.B, Andersen, K.R, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2017-04-05 | | Release date: | 2017-06-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Introducing site-specific cysteines into nanobodies for mercury labelling allows de novo phasing of their crystal structures.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5NLU
 
 | | Structure of Nb36 crystal form 1 | | Descriptor: | SULFATE ION, single domain llama antibody Nb36 | | Authors: | Hansen, S.B, Andersen, K.R, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-31 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.193 Å) | | Cite: | Introducing site-specific cysteines into nanobodies for mercury labelling allows de novo phasing of their crystal structures.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
3GUA
 
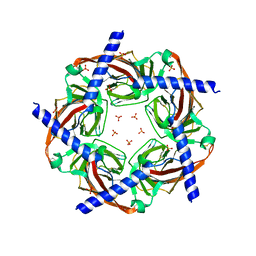 | | Sulfates bound in the vestibule of AChBP | | Descriptor: | SULFATE ION, Soluble acetylcholine receptor | | Authors: | Hansen, S.B, Taylor, P. | | Deposit date: | 2009-03-28 | | Release date: | 2009-07-14 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An Ion Selectivity Filter in the Extracellular Domain of Cys-loop Receptors Reveals Determinants for Ion Conductance
J.Biol.Chem., 283, 2008
|
|
8PEH
 
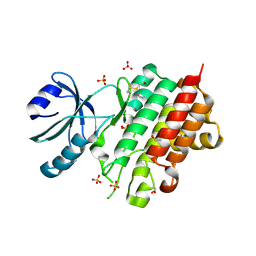 | | Crystal structure of Lotus japonicus SYMRK kinase domain D738N | | Descriptor: | 1,2-ETHANEDIOL, Receptor-like kinase SYMRK, SULFATE ION | | Authors: | Noergaard, M.M.M, Gysel, K, Hansen, S.B, Andersen, K.R. | | Deposit date: | 2023-06-14 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Phosphorylation of the alpha-I motif in SYMRK drives root nodule organogenesis.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
1YI5
 
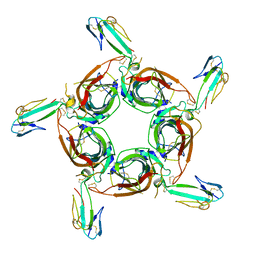 | | Crystal structure of the a-cobratoxin-AChBP complex | | Descriptor: | Acetylcholine-binding protein, Long neurotoxin 1 | | Authors: | Bourne, Y, Talley, T.T, Hansen, S.B, Taylor, P, Marchot, P. | | Deposit date: | 2005-01-11 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Crystal structure of a Cbtx-AChBP complex reveals essential interactions between snake alpha-neurotoxins and nicotinic receptors
Embo J., 24, 2005
|
|
6ZHH
 
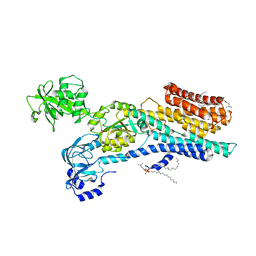 | | Ca2+-ATPase from Listeria Monocytogenes with G4 insertion. | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BERYLLIUM TRIFLUORIDE ION, Calcium-transporting ATPase, ... | | Authors: | Basse Hansen, S, Dyla, M, Neumann, C, Quistgaard, E.M.H, Lauwring Andersen, J, Kjaergaard, M, Nissen, P. | | Deposit date: | 2020-06-23 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Crystal Structure of the Ca 2+ -ATPase 1 from Listeria monocytogenes reveals a Pump Primed for Dephosphorylation.
J.Mol.Biol., 433, 2021
|
|
6ZHF
 
 | | Calcium ATPase-1 from Listeria monocytogenes in complex with BeF | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Calcium-transporting ATPase, MAGNESIUM ION | | Authors: | Basse Hansen, S, Dyla, M, Neumann, C, Quistgaard, E.M.H, Lauwring Andersen, J, Kjaergaard, M, Nissen, P. | | Deposit date: | 2020-06-23 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Crystal Structure of the Ca 2+ -ATPase 1 from Listeria monocytogenes reveals a Pump Primed for Dephosphorylation.
J.Mol.Biol., 433, 2021
|
|
6ZHG
 
 | | Ca2+-ATPase from Listeria Monocytogenes in complex with AlF | | Descriptor: | Calcium-transporting ATPase, MAGNESIUM ION, TETRAFLUOROALUMINATE ION | | Authors: | Basse Hansen, S, Dyla, M, Neumann, C, Quistgaard, E.M.H, Lauwring Andersen, J, Kjaergaard, M, Nissen, P. | | Deposit date: | 2020-06-23 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Crystal Structure of the Ca 2+ -ATPase 1 from Listeria monocytogenes reveals a Pump Primed for Dephosphorylation.
J.Mol.Biol., 433, 2021
|
|
6XWE
 
 | | Crystal structure of LYK3 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETONITRILE, LysM domain receptor-like kinase 3, ... | | Authors: | Gysel, K, Blaise, M, Andersen, K.R. | | Deposit date: | 2020-01-23 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Ligand-recognizing motifs in plant LysM receptors are major determinants of specificity.
Science, 369, 2020
|
|
