1B2T
 
 | |
1DON
 
 | |
1DOM
 
 | |
1C3T
 
 | |
4RWS
 
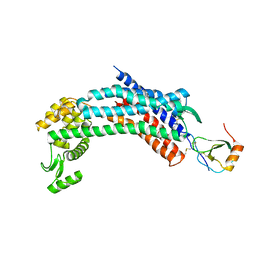 | | Crystal structure of CXCR4 and viral chemokine antagonist vMIP-II complex (PSI Community Target) | | Descriptor: | C-X-C chemokine receptor type 4/Endolysin chimeric protein, Viral macrophage inflammatory protein 2 | | Authors: | Qin, L, Kufareva, I, Holden, L, Wang, C, Zheng, Y, Wu, H, Fenalti, G, Han, G.W, Cherezov, V, Abagyan, R, Stevens, R.C, Handel, T.M, GPCR Network (GPCR) | | Deposit date: | 2014-12-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural biology. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine.
Science, 347, 2015
|
|
1UD7
 
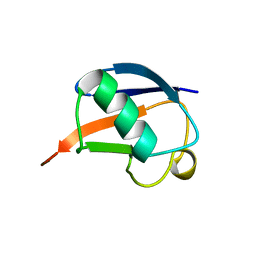 | | SOLUTION STRUCTURE OF THE DESIGNED HYDROPHOBIC CORE MUTANT OF UBIQUITIN, 1D7 | | Descriptor: | PROTEIN (UBIQUITIN CORE MUTANT 1D7) | | Authors: | Johnson, E.C, Lazar, G.A, Desjarlais, J.R, Handel, T.M. | | Deposit date: | 1999-04-07 | | Release date: | 1999-05-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of a designed hydrophobic core variant of ubiquitin.
Structure Fold.Des., 7, 1999
|
|
1DOK
 
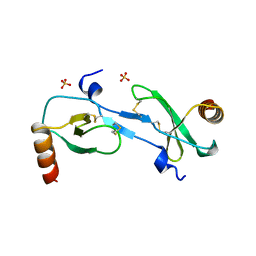 | | MONOCYTE CHEMOATTRACTANT PROTEIN 1, P-FORM | | Descriptor: | MONOCYTE CHEMOATTRACTANT PROTEIN 1, SULFATE ION | | Authors: | Lubkowski, J, Bujacz, G, Boque, L, Wlodawer, A, Domaille, P.J, Handel, T.M. | | Deposit date: | 1996-11-27 | | Release date: | 1997-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of MCP-1 in two crystal forms provides a rare example of variable quaternary interactions.
Nat.Struct.Biol., 4, 1997
|
|
7SK9
 
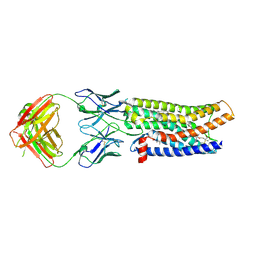 | | Cryo-EM structure of human ACKR3 in complex with a small molecule partial agonist CCX662, and an intracellular Fab | | Descriptor: | (1R)-4-[7-(3-carboxypropoxy)-6-methylquinolin-8-yl]-1-{[2-(4-hydroxypiperidin-1-yl)-1,3-thiazol-4-yl]methyl}-1,4-diazepan-1-ium, Atypical chemokine receptor 3, CHOLESTEROL, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK3
 
 | | Cryo-EM structure of ACKR3 in complex with CXCL12, an intracellular Fab, and an extracellular Fab | | Descriptor: | Atypical chemokine receptor 3, CHOLESTEROL, CID24 Fab heavy chain, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK7
 
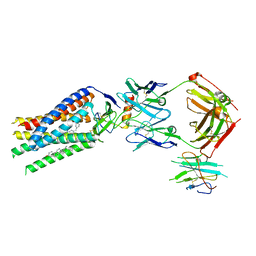 | | Cryo-EM structure of human ACKR3 in complex with CXCL12, a small molecule partial agonist CCX662, and an extracellular Fab | | Descriptor: | (1R)-4-[7-(3-carboxypropoxy)-6-methylquinolin-8-yl]-1-{[2-(4-hydroxypiperidin-1-yl)-1,3-thiazol-4-yl]methyl}-1,4-diazepan-1-ium, Anti-Fab nanobody, Atypical chemokine receptor 3, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK8
 
 | | Cryo-EM structure of human ACKR3 in complex with CXCL12, a small molecule partial agonist CCX662, an extracellular Fab, and an intracellular Fab | | Descriptor: | (1R)-4-[7-(3-carboxypropoxy)-6-methylquinolin-8-yl]-1-{[2-(4-hydroxypiperidin-1-yl)-1,3-thiazol-4-yl]methyl}-1,4-diazepan-1-ium, Atypical chemokine receptor 3, CHOLESTEROL, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK5
 
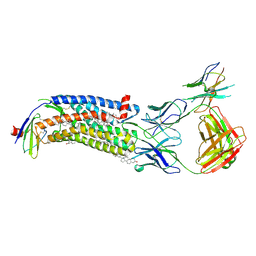 | | Cryo-EM structure of ACKR3 in complex with CXCL12 and an intracellular Fab | | Descriptor: | Anti-Fab nanobody, Atypical chemokine receptor 3, CHOLESTEROL, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK4
 
 | | Cryo-EM structure of ACKR3 in complex with chemokine N-terminal mutant CXCL12_LRHQ, an intracellular Fab, and an extracellular Fab | | Descriptor: | Atypical chemokine receptor 3, CHOLESTEROL, CID24 Fab heavy chain, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
7SK6
 
 | | Cryo-EM structure of human ACKR3 in complex with chemokine N-terminal mutant CXCL12_LRHQ and an intracellular Fab | | Descriptor: | Atypical chemokine receptor 3, CID24 Fab heavy chain, CID24 Fab light chain, ... | | Authors: | Yen, Y.C, Schafer, C.T, Gustavsson, M, Handel, T.M, Tesmer, J.J.G. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of atypical chemokine receptor 3 reveal the basis for its promiscuity and signaling bias.
Sci Adv, 8, 2022
|
|
3C4O
 
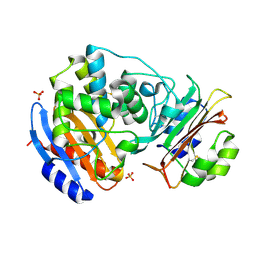 | | Crystal Structure of the SHV-1 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) E73M/S130K/S146M complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein, SULFATE ION | | Authors: | Reynolds, K.A, Hanes, M.S, Thomson, J.M, Antczak, A.J, Berger, J.M, Bonomo, R.A, Kirsch, J.F, Handel, T.M. | | Deposit date: | 2008-01-30 | | Release date: | 2008-05-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational redesign of the SHV-1 beta-lactamase/beta-lactamase inhibitor protein interface.
J.Mol.Biol., 382, 2008
|
|
3C4P
 
 | | Crystal Structure of the SHV-1 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) E73M complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein, SULFATE ION | | Authors: | Reynolds, K.A, Hanes, M.S, Thomson, J.M, Antczak, A.J, Berger, J.M, Bonomo, R.A, Kirsch, J.F, Handel, T.M. | | Deposit date: | 2008-01-30 | | Release date: | 2008-05-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Computational redesign of the SHV-1 beta-lactamase/beta-lactamase inhibitor protein interface.
J.Mol.Biol., 382, 2008
|
|
3E2L
 
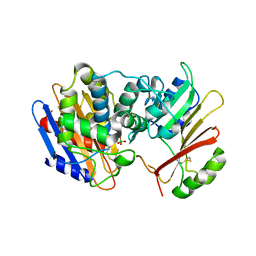 | | Crystal Structure of the KPC-2 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) | | Descriptor: | Beta-lactamase inhibitory protein, Carbapenemase | | Authors: | Hanes, M.S, Jude, K.M, Berger, J.M, Kirsch, J.F, Bonomo, R.A, Handel, T.M. | | Deposit date: | 2008-08-05 | | Release date: | 2009-08-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and biochemical characterization of the interaction between KPC-2 beta-lactamase and beta-lactamase inhibitor protein
Biochemistry, 48, 2009
|
|
3E2K
 
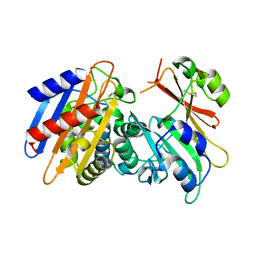 | | Crystal Structure of the KPC-2 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) | | Descriptor: | Beta-lactamase inhibitory protein, Carbapenemase | | Authors: | Hanes, M.S, Jude, K.M, Berger, J.M, Bonomo, R.A, Handel, T.M. | | Deposit date: | 2008-08-05 | | Release date: | 2009-08-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical characterization of the interaction between KPC-2 beta-lactamase and beta-lactamase inhibitor protein
Biochemistry, 48, 2009
|
|
1F2L
 
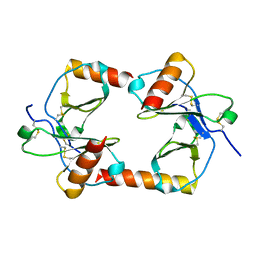 | |
5T1A
 
 | | Structure of CC Chemokine Receptor 2 with Orthosteric and Allosteric Antagonists | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{R})-1-(4-chloranyl-2-fluoranyl-phenyl)-2-cyclohexyl-3-ethanoyl-4-oxidanyl-2~{H}-pyrrol-5-one, (3S)-1-{(1S,2R,4R)-4-[methyl(propan-2-yl)amino]-2-propylcyclohexyl}-3-{[6-(trifluoromethyl)quinazolin-4-yl]amino}pyrrolidin-2-one, ... | | Authors: | Zheng, Y, Qin, L, Ortiz Zacarias, N.V, de Vries, H, Han, G.W, Gustavsson, M, Dabros, M, Zhao, C, Cherney, R.J, Carter, P, Stamos, D, Abagyan, R, Cherezov, V, Stevens, R.C, IJzerman, A.P, Heitman, L.H, Tebben, A, Kufareva, I, Handel, T.M. | | Deposit date: | 2016-08-18 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists.
Nature, 540, 2016
|
|
5UIW
 
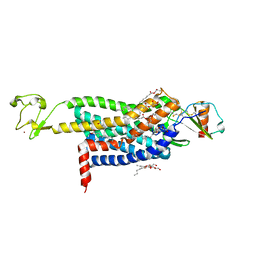 | | Crystal Structure of CC Chemokine Receptor 5 (CCR5) in complex with high potency HIV entry inhibitor 5P7-CCL5 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, C-C chemokine receptor type 5,Rubredoxin chimera, C-C motif chemokine 5, ... | | Authors: | Zheng, Y, Qin, L, Han, G.W, Gustavsson, M, Kawamura, T, Stevens, R.C, Cherezov, V, Kufareva, I, Handel, T.M. | | Deposit date: | 2017-01-15 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structure of CC Chemokine Receptor 5 with a Potent Chemokine Antagonist Reveals Mechanisms of Chemokine Recognition and Molecular Mimicry by HIV.
Immunity, 46, 2017
|
|
2G2W
 
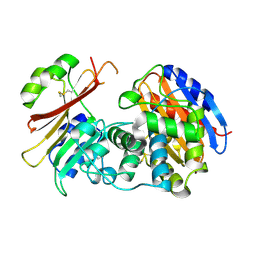 | | Crystal Structure of the SHV D104K Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein | | Authors: | Reynolds, K.A, Thomson, J.M, Corbett, K.D, Bethel, C.R, Berger, J.M, Kirsch, J.F, Bonomo, R.A, Handel, T.M. | | Deposit date: | 2006-02-16 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Computational Characterization of the SHV-1 beta-Lactamase-beta-Lactamase Inhibitor Protein Interface.
J.Biol.Chem., 281, 2006
|
|
2G2U
 
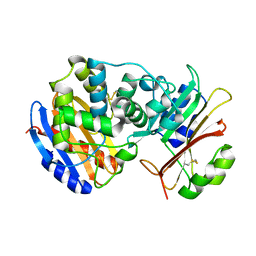 | | Crystal Structure of the SHV-1 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein | | Authors: | Reynolds, K.A, Thomson, J.M, Corbett, K.D, Bethel, C.R, Berger, J.M, Kirsch, J.F, Bonomo, R.A, Handel, T.M. | | Deposit date: | 2006-02-16 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Computational Characterization of the SHV-1 beta-Lactamase-beta-Lactamase Inhibitor Protein Interface.
J.Biol.Chem., 281, 2006
|
|
2KUM
 
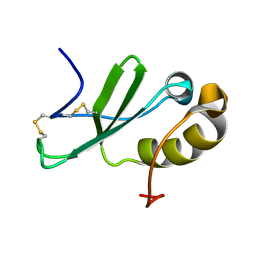 | | Solution structure of the human chemokine CCL27 | | Descriptor: | C-C motif chemokine 27 | | Authors: | Kirkpatrick, J.P, Jansma, A, Hsu, A, Handel, T.M, Nietlispach, D. | | Deposit date: | 2010-02-22 | | Release date: | 2010-03-02 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | NMR analysis of the structure, dynamics, and unique oligomerization properties of the chemokine CCL27.
J.Biol.Chem., 285, 2010
|
|
2L9H
 
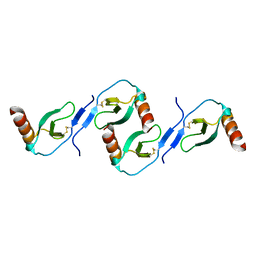 | | Oligomeric Structure of the Chemokine CCL5/RANTES from NMR, MS, and SAXS Data | | Descriptor: | C-C motif chemokine 5 | | Authors: | Wang, X, Watson, C.M, Sharp, J.S, Handel, T.M, Prestegard, J.H. | | Deposit date: | 2011-02-09 | | Release date: | 2011-06-22 | | Last modified: | 2011-08-24 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Oligomeric Structure of the Chemokine CCL5/RANTES from NMR, MS, and SAXS Data.
Structure, 19, 2011
|
|
