6KM7
 
 | | The structural basis for the internal interaction in RBBP5 | | Descriptor: | HEXAETHYLENE GLYCOL, NONAETHYLENE GLYCOL, Retinoblastoma-binding protein 5, ... | | Authors: | Han, J, Li, T, Chen, Y. | | Deposit date: | 2019-07-31 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The internal interaction in RBBP5 regulates assembly and activity of MLL1 methyltransferase complex.
Nucleic Acids Res., 47, 2019
|
|
7MEM
 
 | |
7T3D
 
 | | CryoEM map of anchor 222-1C06 Fab and lateral patch 2B05 Fab binding H1 HA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 222-1C06 mAb heavy chain, ... | | Authors: | Han, J, Ward, A.B. | | Deposit date: | 2021-12-07 | | Release date: | 2022-01-12 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Broadly neutralizing antibodies target a haemagglutinin anchor epitope.
Nature, 602, 2022
|
|
6KZ7
 
 | | The crystal structure of BAF155 SWIRM domain and N-terminal elongated hSNF5 RPT1 domain complex: Chromatin remodeling complex | | Descriptor: | SWI/SNF complex subunit SMARCC1, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | Authors: | Lee, W, Han, J, Kim, I, Park, J.H, Joo, K, Lee, J, Suh, J.Y. | | Deposit date: | 2019-09-23 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A Coil-to-Helix Transition Serves as a Binding Motif for hSNF5 and BAF155 Interaction.
Int J Mol Sci, 21, 2020
|
|
6LZP
 
 | |
7MU8
 
 | | Structure of the minimally glycosylated human CEACAM1 N-terminal domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carcinoembryonic antigen-related cell adhesion molecule 1, GLYCEROL, ... | | Authors: | Belcher Dufrisne, M, Swope, N, Kieber, M, Yang, J.Y, Han, J, Li, J, Moremen, K.W, Prestegard, J.H, Columbus, L. | | Deposit date: | 2021-05-14 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Human CEACAM1 N-domain dimerization is independent from glycan modifications.
Structure, 30, 2022
|
|
1LJT
 
 | | Crystal Structure of Macrophage Migration Inhibitory Factor complexed with (S,R)-3-(4-hydroxyphenyl)-4,5-dihydro-5-isoxazole-acetic acid methyl ester (ISO-1) | | Descriptor: | 3-(4-HYDROXYPHENYL)-4,5-DIHYDRO-5-ISOXAZOLE-ACETIC ACID METHYL ESTER, Macrophage migration inhibitory factor | | Authors: | Lubetsky, J.B, Dios, A, Han, J, Aljabari, B, Ruzsicska, B, Mitchell, R, Lolis, E, Al-Abed, Y. | | Deposit date: | 2002-04-22 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Tautomerase Active Site of Macrophage Migration Inhibitory factor is a Potential Target for Discovery of Novel Anti-inflammatory Agents
J.Biol.Chem., 277, 2002
|
|
8TP7
 
 | | H2 hemagglutinin (A/Singapore/1/1957) in complex with Sa-targeting Fab 4-1-1G03 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of monoclonal antibody 4-1-1G03, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
8TP4
 
 | | H2 hemagglutinin (A/Singapore/1/1957) in complex with RBS-targeting Fab 1-1-1E04 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 1-1-1E04, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
8TPA
 
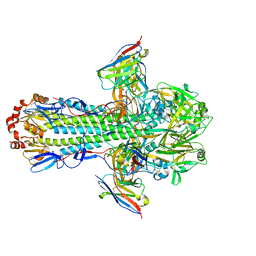 | | H1 hemagglutinin (NC99) in complex with medial-junction-targeting Fab 2-2-1G06 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 2-2-1G06, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
8TP6
 
 | | H2 hemagglutinin (A/Singapore/1/1957) in complex with RBS-targeting Fab 4-1-1E02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 4-1-1E02 Fab, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
8TP3
 
 | | H1 hemagglutinin (NC99) in complex with RBS-targeting Fab 1-1-1F05 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 1-1-1F05, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
8TP2
 
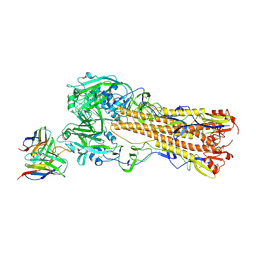 | | H2 hemagglutinin (A/Singapore/1/1957) in complex with RBS-targeting 1-1-1F05 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 1-1-1F05, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
8TP9
 
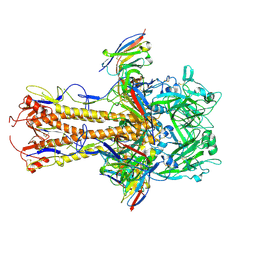 | | H2 hemagglutinin (A/Singapore/1/1957) in complex with medial-junction-targeting Fab 2-2-1G06 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 2-2-1G06, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
8TP5
 
 | | H1 hemagglutinin (NC99) in complex with RBS-targeting Fab 1-1-1E04 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 1-1-1E04, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
5EJO
 
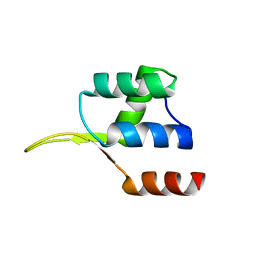 | | Crystal structure of the winged helix domain in Chromatin assembly factor 1 subunit p90 | | Descriptor: | Chromatin assembly factor 1 subunit p90 | | Authors: | Zhang, K, Gao, Y, Li, J, Burgess, R, Han, J, Liang, H, Zhang, Z, Liu, Y. | | Deposit date: | 2015-11-02 | | Release date: | 2016-03-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A DNA binding winged helix domain in CAF-1 functions with PCNA to stabilize CAF-1 at replication forks
Nucleic Acids Res., 44, 2016
|
|
6KY5
 
 | | Crystal structure of a hydrolase mutant | | Descriptor: | PET hydrolase, SULFATE ION | | Authors: | Cui, Y.L, Chen, Y.C, Liu, X.Y, Dong, S.J, Han, J, Xiang, H, Chen, Q, Liu, H.Y, Han, X, Liu, W.D, Tang, S.Y, Wu, B. | | Deposit date: | 2019-09-16 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Computational redesign of PETase for plasticbiodegradation by GRAPE strategy.
Biorxiv, 2020
|
|
6JCG
 
 | | Room temperature structure of HIV-1 Integrase catalytic core domain by serial femtosecond crystallography. | | Descriptor: | CACODYLATE ION, Integrase | | Authors: | Park, J.H, Shi, Y, Han, J, Li, X, Kim, T.H, Yun, J.H. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-Cryogenic Structure and Dynamics of HIV-1 Integrase Catalytic Core Domain by X-ray Free-Electron Lasers.
Int J Mol Sci, 20, 2019
|
|
6JCF
 
 | | Cryogenic structure of HIV-1 Integrase catalytic core domain by synchrotron | | Descriptor: | CACODYLATE ION, Integrase | | Authors: | Park, J.H, Han, J, Kim, T.H, Yun, J.H, Lee, W. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Non-Cryogenic Structure and Dynamics of HIV-1 Integrase Catalytic Core Domain by X-ray Free-Electron Lasers.
Int J Mol Sci, 20, 2019
|
|
8DZR
 
 | | GR89,696 bound Kappa Opioid Receptor in complex with gustducin | | Descriptor: | G alpha gustducin protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Fay, J.F, Che, T. | | Deposit date: | 2022-08-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Ligand and G-protein selectivity in the kappa-opioid receptor.
Nature, 617, 2023
|
|
8DZQ
 
 | | momSalB bound Kappa Opioid Receptor in complex with GoA | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(o) subunit alpha, ... | | Authors: | Fay, J.F, Che, T. | | Deposit date: | 2022-08-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Ligand and G-protein selectivity in the kappa-opioid receptor.
Nature, 617, 2023
|
|
8DZS
 
 | | GR89,696 bound Kappa Opioid Receptor in complex with Gz | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(z) subunit alpha, ... | | Authors: | Fay, J.F, Che, T. | | Deposit date: | 2022-08-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Ligand and G-protein selectivity in the kappa-opioid receptor.
Nature, 617, 2023
|
|
8DZP
 
 | | momSalB bound Kappa Opioid Receptor in complex with Gi1 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Fay, J.F, Che, T. | | Deposit date: | 2022-08-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Ligand and G-protein selectivity in the kappa-opioid receptor.
Nature, 617, 2023
|
|
3UL8
 
 | | Crystal structure of the TV3 mutant V134L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Toll-like receptor 4, ... | | Authors: | Kim, H.J, Cheong, H.K, Jeon, Y.H. | | Deposit date: | 2011-11-10 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Rational Design of a Toll-like Receptor 4 (TLR4) Decoy Receptor with High Binding Affinity for a Target Protein.
Plos One, 7, 2012
|
|
3UL7
 
 | | Crystal structure of the TV3 mutant F63W | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Toll-like receptor 4, ... | | Authors: | Kim, H.J, Cheong, H.K, Jeon, Y.H. | | Deposit date: | 2011-11-10 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-Based Rational Design of a Toll-like Receptor 4 (TLR4) Decoy Receptor with High Binding Affinity for a Target Protein.
Plos One, 7, 2012
|
|
