2MBS
 
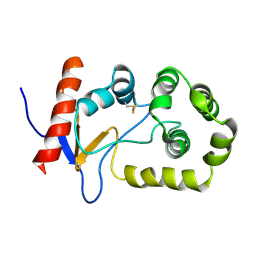 | | NMR solution structure of oxidized KpDsbA | | Descriptor: | Thiol:disulfide interchange protein | | Authors: | Kurth, F, Rimmer, K, Premkumar, L, Mohanty, B, Duprez, W, Halili, M.A, Shouldice, S.R, Heras, B, Fairlie, D.P, Scanlon, M.J, Martin, J.L. | | Deposit date: | 2013-08-03 | | Release date: | 2013-12-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Comparative Sequence, Structure and Redox Analyses of Klebsiella pneumoniae DsbA Show That Anti-Virulence Target DsbA Enzymes Fall into Distinct Classes.
Plos One, 8, 2013
|
|
4XVW
 
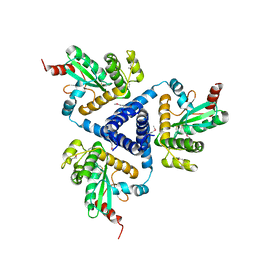 | |
5ID4
 
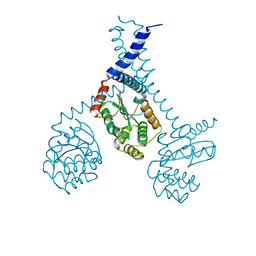 | |
5IDR
 
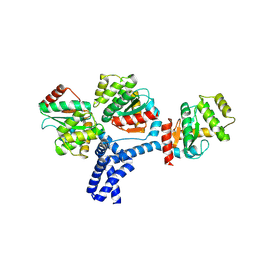 | |
4TKY
 
 | |
7RGV
 
 | |
6EEZ
 
 | |
4DVC
 
 | | Structural and functional studies of TcpG, the Vibrio cholerae DsbA disulfide-forming protein required for pilus and cholera toxin production | | Descriptor: | DIMETHYL SULFOXIDE, SULFATE ION, Thiol:disulfide interchange protein DsbA | | Authors: | Walden, P.M, Martin, J.L. | | Deposit date: | 2012-02-23 | | Release date: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2 A resolution crystal structure of TcpG, the Vibrio cholerae DsbA disulfide-forming protein required for pilus and cholera-toxin production
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4MCU
 
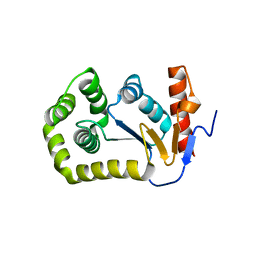 | |
4P3Y
 
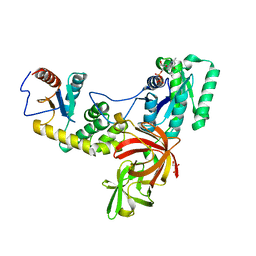 | |
7M24
 
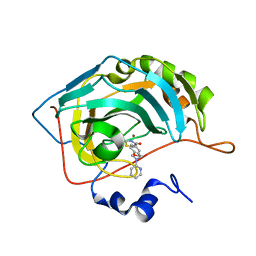 | |
7LUJ
 
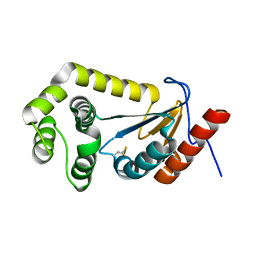 | |
7M26
 
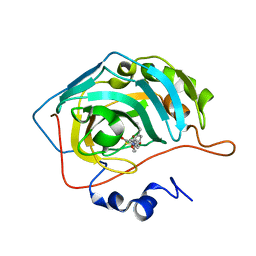 | | Human carbonic anhydrase II in complex with pioglitazone | | Descriptor: | (5R)-5-{4-[2-(5-ethylpyridin-2-yl)ethoxy]benzyl}-1,3-thiazolidine-2,4-dione, Carbonic anhydrase 2, ZINC ION | | Authors: | Mueller, S.L, Peat, T.S. | | Deposit date: | 2021-03-16 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Glitazone Class of Drugs as Carbonic Anhydrase Inhibitors-A Spin-Off Discovery from Fragment Screening.
Molecules, 26, 2021
|
|
7M23
 
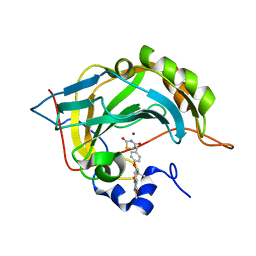 | | Human carbonic anhydrase II in complex with troglitazone | | Descriptor: | (5R)-5-(4-{[(2R)-6-HYDROXY-2,5,7,8-TETRAMETHYL-3,4-DIHYDRO-2H-CHROMEN-2-YL]METHOXY}BENZYL)-1,3-THIAZOLIDINE-2,4-DIONE, Carbonic anhydrase 2, ZINC ION | | Authors: | Mueller, S.L, Peat, T.S. | | Deposit date: | 2021-03-16 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Glitazone Class of Drugs as Carbonic Anhydrase Inhibitors-A Spin-Off Discovery from Fragment Screening.
Molecules, 26, 2021
|
|
7LUH
 
 | |
