3NDB
 
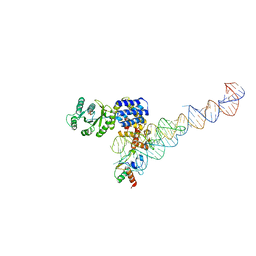 | | Crystal structure of a signal sequence bound to the signal recognition particle | | Descriptor: | PHOSPHATE ION, SRP RNA, Signal recognition 54 kDa protein, ... | | Authors: | Hainzl, T, Huang, S, Sauer-Eriksson, E. | | Deposit date: | 2010-06-07 | | Release date: | 2011-02-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of signal-sequence recognition by the signal recognition particle.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4XCO
 
 | |
1LNG
 
 | |
2V3C
 
 | |
1Z43
 
 | |
8C7O
 
 | |
8C7U
 
 | |
8C7T
 
 | |
8C7S
 
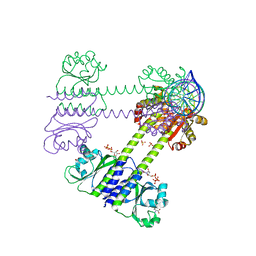 | | Transcriptional pleiotropic repressor CodY from Staphylococcus aureus in complex with Ile, GTP, and a 30-bp DNA fragment encompassing two overlapping binding sites | | Descriptor: | DNA (30-MER), GUANOSINE-5'-TRIPHOSPHATE, Global transcriptional regulator CodY (Fragment), ... | | Authors: | Hainzl, T, Sauer-Eriksson, A.E. | | Deposit date: | 2023-01-17 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural insights into CodY activation and DNA recognition.
Nucleic Acids Res., 51, 2023
|
|
4XIX
 
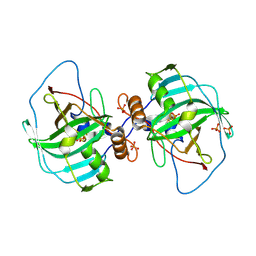 | | Carbonic anhydrase Cah3 from Chlamydomonas reinhardtii in complex with phosphate. | | Descriptor: | Carbonic anhydrase, alpha type, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Hainzl, T, Grundstrom, C, Benlloch, R, Shevela, D, Shutova, T, Messinger, J, Samuelsson, G, Sauer-Eriksson, A.E. | | Deposit date: | 2015-01-08 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure and Functional Characterization of Photosystem II-Associated Carbonic Anhydrase CAH3 in Chlamydomonas reinhardtii.
Plant Physiol., 167, 2015
|
|
4XIW
 
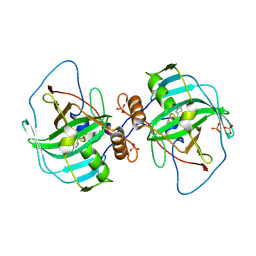 | | Carbonic anhydrase Cah3 from Chlamydomonas reinhardtii in complex with acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase, alpha type, ... | | Authors: | Hainzl, T, Grundstrom, C, Benlloch, R, Shevela, D, Shutova, T, Messinger, J, Samuelsson, G, Sauer-Eriksson, A.E. | | Deposit date: | 2015-01-07 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure and Functional Characterization of Photosystem II-Associated Carbonic Anhydrase CAH3 in Chlamydomonas reinhardtii.
Plant Physiol., 167, 2015
|
|
1DUH
 
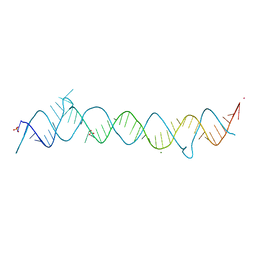 | | CRYSTAL STRUCTURE OF THE CONSERVED DOMAIN IV OF E. COLI 4.5S RNA | | Descriptor: | 4.5S RNA DOMAIN IV, LUTETIUM (III) ION, MAGNESIUM ION, ... | | Authors: | Jovine, L, Hainzl, T, Oubridge, C, Scott, W.G, Li, J, Sixma, T.K, Wonacott, A, Skarzynski, T, Nagai, K. | | Deposit date: | 2000-01-17 | | Release date: | 2000-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the ffh and EF-G binding sites in the conserved domain IV of Escherichia coli 4.5S RNA.
Structure Fold.Des., 8, 2000
|
|
3UCM
 
 | |
3UCK
 
 | | Coccomyxa beta-carbonic anhydrase in complex with phosphate | | Descriptor: | CHLORIDE ION, Carbonic anhydrase, PHOSPHATE ION, ... | | Authors: | Huang, S, Hainzl, T, Sauer-Eriksson, A.E. | | Deposit date: | 2011-10-27 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies of [beta]-carbonic anhydrase from the green alga Coccomyxa: inhibitor complexes with anions and acetazolamide.
Plos One, 6, 2011
|
|
3UCO
 
 | |
3UCN
 
 | | Coccomyxa beta-carbonic anhydrase in complex with azide | | Descriptor: | AZIDE ION, CHLORIDE ION, Carbonic anhydrase, ... | | Authors: | Huang, S, Hainzl, T, Sauer-Eriksson, A.E. | | Deposit date: | 2011-10-27 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural studies of [beta]-carbonic anhydrase from the green alga Coccomyxa: inhibitor complexes with anions and acetazolamide.
Plos One, 6, 2011
|
|
3UCJ
 
 | | Coccomyxa beta-carbonic anhydrase in complex with acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CHLORIDE ION, Carbonic anhydrase, ... | | Authors: | Huang, S, Hainzl, T, Sauer-Eriksson, A.E. | | Deposit date: | 2011-10-27 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies of [beta]-carbonic anhydrase from the green alga Coccomyxa: inhibitor complexes with anions and acetazolamide.
Plos One, 6, 2011
|
|
