1NLY
 
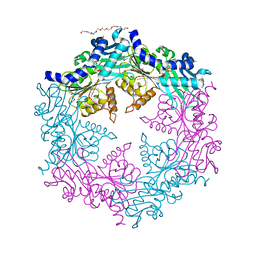 | | Crystal structure of the traffic ATPase of the Helicobacter pylori type IV secretion system in complex with ATPgammaS | | Descriptor: | MAGNESIUM ION, NONAETHYLENE GLYCOL, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Savvides, S.N, Yeo, H.J, Beck, M.R, Blaesing, F, Lurz, R, Lanka, E, Buhrdorf, R, Fischer, W, Haas, R, Waksman, G. | | Deposit date: | 2003-01-08 | | Release date: | 2003-05-06 | | Last modified: | 2012-03-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | VirB11 ATPases are dynamic hexameric assemblies: New insights into bacterial type IV secretion
Embo J., 22, 2003
|
|
2PT7
 
 | | Crystal structure of Cag VirB11 (HP0525) and an inhibitory protein (HP1451) | | Descriptor: | Cag-alfa, Hypothetical protein | | Authors: | Hare, S, Fischer, W, Williams, R, Terradot, L, Bayliss, R, Haas, R, Waksman, G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification, structure and mode of action of a new regulator of the Helicobacter pylori HP0525 ATPase.
Embo J., 26, 2007
|
|
1NLZ
 
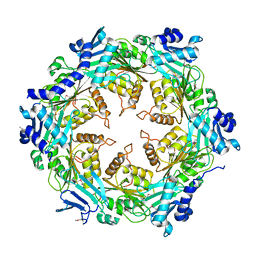 | | Crystal structure of unliganded traffic ATPase of the type IV secretion system of helicobacter pylori | | Descriptor: | virB11 homolog | | Authors: | Savvides, S.N, Yeo, H.J, Beck, M.R, Blaesing, F, Lurz, R, Lanka, E, Buhrdorf, R, Fischer, W, Haas, R, Waksman, G. | | Deposit date: | 2003-01-08 | | Release date: | 2003-05-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | VirB11 ATPases are dynamic hexameric assemblies: New insights into bacterial type IV secretion
Embo J., 22, 2003
|
|
1OPX
 
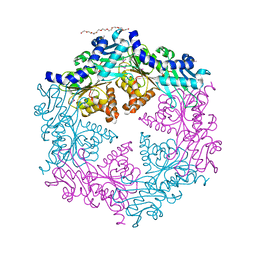 | | Crystal structure of the traffic ATPase (HP0525) of the Helicobacter pylori type IV secretion system bound by sulfate | | Descriptor: | NONAETHYLENE GLYCOL, SULFATE ION, virB11 homolog | | Authors: | Savvides, S.N, Yeo, H.J, Beck, M.R, Blaesing, F, Lurz, R, Lanka, E, Buhrdorf, R, Fischer, W, Haas, R, Waksman, G. | | Deposit date: | 2003-03-06 | | Release date: | 2003-05-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | VirB11 ATPases are dynamic hexameric assemblies: New insights into bacterial type IV secretion
Embo J., 22, 2003
|
|
1QWL
 
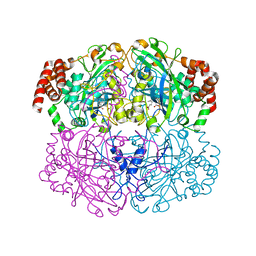 | | Structure of Helicobacter pylori catalase | | Descriptor: | AZIDE ION, KatA catalase, OXYGEN MOLECULE, ... | | Authors: | Loewen, P.C, Carpena, X, Perez-Luque, R, Rovira, C, Haas, R, Obenbreit, S, Nicholls, P, Fita, I. | | Deposit date: | 2003-09-02 | | Release date: | 2004-03-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Helicobacter pylori Catalase, with and without Formic Acid Bound, at 1.6 A Resolution
Biochemistry, 43, 2004
|
|
1QWM
 
 | | Structure of Helicobacter pylori catalase with formic acid bound | | Descriptor: | AZIDE ION, FORMIC ACID, KatA catalase, ... | | Authors: | Loewen, P.C, Carpena, X, Perez-Luque, R, Rovira, C, Haas, R, Odenbreit, S, Nicholls, P, Fita, I. | | Deposit date: | 2003-09-02 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Helicobacter pylori Catalase, with and without Formic Acid Bound, at 1.6 A Resolution
Biochemistry, 43, 2004
|
|
5LP2
 
 | |
2IDM
 
 | | 2.00 A Structure of T87I/Y106W Phosphono-CheY | | Descriptor: | ACETATE ION, Chemotaxis protein cheY | | Authors: | Halkides, C.J, Haas, R.M, McAdams, K.A, Casper, E.S, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2006-09-15 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY help to explain their binding affinities to the FliM and CheZ peptides.
Arch.Biochem.Biophys., 479, 2008
|
|
2ID7
 
 | | 1.75 A Structure of T87I Phosphono-CheY | | Descriptor: | Chemotaxis protein cheY | | Authors: | Halkides, C.J, Haas, R.M, McAdams, K.A, Casper, E.S, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2006-09-14 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY help to explain their binding affinities to the FliM and CheZ peptides.
Arch.Biochem.Biophys., 479, 2008
|
|
2ID9
 
 | | 1.85 A Structure of T87I/Y106W Phosphono-CheY | | Descriptor: | Chemotaxis protein cheY | | Authors: | Halkides, C.J, Haas, R.M, McAdams, K.A, Casper, E.S, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2006-09-14 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY help to explain their binding affinities to the FliM and CheZ peptides.
Arch.Biochem.Biophys., 479, 2008
|
|
4X5U
 
 | | X-ray crystal structure of CagL at pH 4.2 | | Descriptor: | Cag pathogenicity island protein (Cag18) | | Authors: | Sundberg, E.J, Bonsor, D.A, Diederichs, K. | | Deposit date: | 2014-12-05 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Integrin Engagement by the Helical RGD Motif of the Helicobacter pylori CagL Protein Is Regulated by pH-induced Displacement of a Neighboring Helix.
J.Biol.Chem., 290, 2015
|
|
4G0H
 
 | | Crystal structure of the N-terminal domain of Helicobacter pylori CagA protein | | Descriptor: | Cytotoxicity-associated immunodominant antigen | | Authors: | Kaplan-Turkoz, B, Remaut, H, Dian, C, Louche, A, Terradot, L. | | Deposit date: | 2012-07-09 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insights into Helicobacter pylori oncoprotein CagA interaction with beta 1 integrin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6AW2
 
 | | Crystal structure of the HopQ-CEACAM1 complex | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, HopQ | | Authors: | Bonsor, D.A, Sundberg, E.J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | TheHelicobacter pyloriadhesin protein HopQ exploits the dimer interface of human CEACAMs to facilitate translocation of the oncoprotein CagA.
EMBO J., 37, 2018
|
|
6AVZ
 
 | | Crystal structure of the HopQ-CEACAM3 WT complex | | Descriptor: | CALCIUM ION, Carcinoembryonic antigen-related cell adhesion molecule 3, HopQ, ... | | Authors: | Bonsor, D.A, Sundberg, E.J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-05-16 | | Last modified: | 2018-07-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | TheHelicobacter pyloriadhesin protein HopQ exploits the dimer interface of human CEACAMs to facilitate translocation of the oncoprotein CagA.
EMBO J., 37, 2018
|
|
6AW1
 
 | | Crystal structure of CEACAM3 | | Descriptor: | CHLORIDE ION, Carcinoembryonic antigen-related cell adhesion molecule 3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bonsor, D.A, Sundberg, E.J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | TheHelicobacter pyloriadhesin protein HopQ exploits the dimer interface of human CEACAMs to facilitate translocation of the oncoprotein CagA.
EMBO J., 37, 2018
|
|
6AW3
 
 | | Crystal structure of the HopQ-CEACAM3 L44Q complex | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 3, HopQ | | Authors: | Bonsor, D.A, Sundberg, E.J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-05-16 | | Last modified: | 2018-07-11 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | TheHelicobacter pyloriadhesin protein HopQ exploits the dimer interface of human CEACAMs to facilitate translocation of the oncoprotein CagA.
EMBO J., 37, 2018
|
|
6AW0
 
 | | Crystal structure of CEACAM3 L44Q | | Descriptor: | CHLORIDE ION, Carcinoembryonic antigen-related cell adhesion molecule 3, GLYCEROL, ... | | Authors: | Bonsor, D.A, Sundberg, E.J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | TheHelicobacter pyloriadhesin protein HopQ exploits the dimer interface of human CEACAMs to facilitate translocation of the oncoprotein CagA.
EMBO J., 37, 2018
|
|
