4V62
 
 | | Crystal Structure of cyanobacterial Photosystem II | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Guskov, A, Gabdulkhakov, A, Kern, J, Broser, M, Zouni, A, Saenger, W. | | Deposit date: | 2008-01-17 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride
Nat.Struct.Mol.Biol., 16, 2009
|
|
4EGW
 
 | | The structure of the soluble domain of CorA from Methanocaldococcus jannaschii | | Descriptor: | 1,4-BUTANEDIOL, HEXANE-1,6-DIOL, MAGNESIUM ION, ... | | Authors: | Guskov, A, Nordin, N, Reynaud, A, Engman, H, Lundback, A.-K, Jong, A.J.O, Cornvik, T, Phua, T, Eshaghi, S. | | Deposit date: | 2012-04-02 | | Release date: | 2012-10-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the mechanisms of Mg2+ uptake, transport, and gating by CorA
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4EV6
 
 | | The complete structure of CorA magnesium transporter from Methanocaldococcus jannaschii | | Descriptor: | MAGNESIUM ION, Magnesium transport protein CorA, UNDECYL-MALTOSIDE | | Authors: | Guskov, A, Nordin, N, Reynaud, A, Engman, H, Lundback, A.-K, Jong, A.J.O, Cornvik, T, Phua, T, Eshaghi, S. | | Deposit date: | 2012-04-25 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the mechanisms of Mg2+ uptake, transport, and gating by CorA
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5DWY
 
 | |
5E9S
 
 | |
5D3M
 
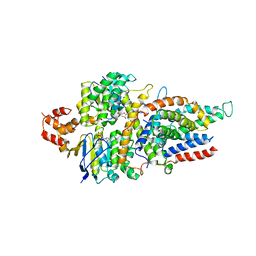 | | Folate ECF transporter: AMPPNP bound state | | Descriptor: | Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, Energy-coupling factor transporter transmembrane protein EcfT, ... | | Authors: | Guskov, A, Swier, L.J.Y.M, Slotboom, D.J. | | Deposit date: | 2015-08-06 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.303 Å) | | Cite: | Structural insight in the toppling mechanism of an energy-coupling factor transporter.
Nat Commun, 7, 2016
|
|
4LA9
 
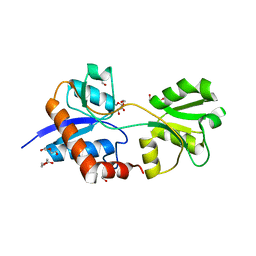 | | Crystal structure of an empty substrate binding domain 1 (SBD1) OF ABC TRANSPORTER GLNPQ from lactococcus lactis | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glutamine ABC transporter permease and substrate binding protein protein, PHOSPHATE ION | | Authors: | Guskov, A, Schuurman-Wolters, G.K, Slotboom, D.J, Poolman, B. | | Deposit date: | 2013-06-19 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Functional Diversity of Tandem Substrate-Binding Domains in ABC Transporters from Pathogenic Bacteria.
Structure, 21, 2013
|
|
4KY0
 
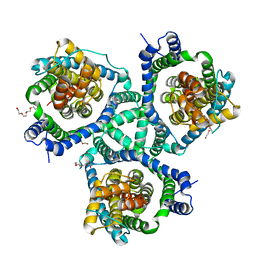 | | Crystal structure of a substrate-free glutamate transporter homologue from Thermococcus kodakarensis | | Descriptor: | Proton/glutamate symporter, SDF family, TETRAETHYLENE GLYCOL | | Authors: | Guskov, A, Jensen, S, Rempel, S, Hanelt, I, Slotboom, D.J. | | Deposit date: | 2013-05-28 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a substrate-free aspartate transporter.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4QTN
 
 | |
6FXG
 
 | |
4V82
 
 | | Crystal structure of cyanobacterial Photosystem II in complex with terbutryn | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Gabdulkhakov, A, Broser, M, Guskov, A, Kern, J, Glockner, C, Muh, F, Saenger, W, Zouni, A. | | Deposit date: | 2010-11-30 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of cyanobacterial photosystem II Inhibition by the herbicide terbutryn
J.Biol.Chem., 286, 2011
|
|
8ODS
 
 | |
3KZI
 
 | | Crystal Structure of Monomeric Form of Cyanobacterial Photosystem II | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Gabdulkhakov, A, Guskov, A, Broser, M, Kern, J, Zouni, A, Saenger, W. | | Deposit date: | 2009-12-08 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal Structure of Monomeric Photosystem II from Thermosynechococcus elongatus at 3.6-A Resolution
J.Biol.Chem., 285, 2010
|
|
8Q5I
 
 | | Structure of Candida albicans 80S ribosome in complex with cephaeline | | Descriptor: | 18S ribosomal RNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Kolosova, O, Zgadzay, Y, Stetsenko, A, Atamas, A, Guskov, A, Yusupov, M. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structural characterization of cephaeline binding to the eukaryotic ribosome using Cryo-Electron Microscopy
Biopolym Cell, 2023
|
|
5JSZ
 
 | | Folate ECF transporter: apo state | | Descriptor: | Conserved hypothetical membrane protein, Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, ... | | Authors: | Swier, L.J.Y.M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2016-05-09 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Structural insight in the toppling mechanism of an energy-coupling factor transporter.
Nat Commun, 7, 2016
|
|
7ZC0
 
 | | 4,6-alpha-glucanotransferase GtfC from Geobacillus 12AMOR1 | | Descriptor: | 4,6-alpha-Glucanotransferase, CALCIUM ION, GLYCEROL | | Authors: | Pijning, T, Dijkhuizen, L, Guskov, A, te Poele, E.M. | | Deposit date: | 2022-03-25 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of 4,6-alpha-Glucanotransferase GtfC-Delta C from Thermophilic Geobacillus 12AMOR1: Starch Transglycosylation in Non-Permuted GH70 Enzymes.
J.Agric.Food Chem., 70, 2022
|
|
7NH9
 
 | |
7FDZ
 
 | | Levansucrase from Brenneria sp. EniD 312 with sucrose | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Xu, W, Hou, X.D, Rao, Y.J, Pijning, T, Guskov, A, Mu, W.M. | | Deposit date: | 2021-07-19 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of Levansucrase from the Gram-Negative Bacterium Brenneria Provides Insights into Its Product Size Specificity.
J.Agric.Food Chem., 70, 2022
|
|
5N9Y
 
 | | The full-length structure of ZntB | | Descriptor: | Zinc transport protein ZntB | | Authors: | Cornelius, G, Stetsenko, A, Scheres, S.H.W, Slotboom, D.J, Guskov, A. | | Deposit date: | 2017-02-27 | | Release date: | 2017-11-15 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural basis of proton driven zinc transport by ZntB.
Nat Commun, 8, 2017
|
|
7NGH
 
 | | Structure of glutamate transporter homologue in complex with Sybody | | Descriptor: | ASPARTIC ACID, Proton/glutamate symporter, SDF family, ... | | Authors: | Arkhipova, V, Slotboom, D.J, Guskov, A. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Kinetic mechanism of Na + -coupled aspartate transport catalyzed by Glt Tk .
Commun Biol, 4, 2021
|
|
5MYJ
 
 | | Structure of 70S ribosome from Lactococcus lactis | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Franken, L.E, Oostergetel, G.T, Pijning, T, Puri, P, Boekema, E.J, Poolman, B, Guskov, A. | | Deposit date: | 2017-01-26 | | Release date: | 2017-10-11 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | A general mechanism of ribosome dimerization revealed by single-particle cryo-electron microscopy.
Nat Commun, 8, 2017
|
|
7EHS
 
 | | Levansucrase from Brenneria sp. EniD 312 | | Descriptor: | GLYCEROL, Levansucrase, NONAETHYLENE GLYCOL, ... | | Authors: | Xu, W, Ni, D.W, Hou, X.D, Rao, Y.J, Pijning, T, Guskov, A, Mu, W.M. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Levansucrase from the Gram-Negative Bacterium Brenneria Provides Insights into Its Product Size Specificity.
J.Agric.Food Chem., 70, 2022
|
|
7EHT
 
 | | Levansucrase from Brenneria sp. EniD 312 | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, GLYCEROL, Levansucrase, ... | | Authors: | Xu, W, Hou, X.D, Rao, Y.J, Pijning, T, Guskov, A, Mu, W.M. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of Levansucrase from the Gram-Negative Bacterium Brenneria Provides Insights into Its Product Size Specificity.
J.Agric.Food Chem., 70, 2022
|
|
7EHR
 
 | | Levansucrase from Brenneria sp. EniD 312 at 1.33 angstroms resolution | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Xu, W, Ni, D.W, Hou, X.D, Rao, Y.J, Pijning, T, Guskov, A, Mu, W.M. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Crystal Structure of Levansucrase from the Gram-Negative Bacterium Brenneria Provides Insights into Its Product Size Specificity.
J.Agric.Food Chem., 70, 2022
|
|
7Q0Q
 
 | | Acetyltrasferase(3) type IIIa in complex with 3-N-methyl-nemycin B | | Descriptor: | 3N methyl nemycin B, ACETATE ION, Aminoglycoside N(3)-acetyltransferase III, ... | | Authors: | Pontillo, N, Guskov, A. | | Deposit date: | 2021-10-16 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The 3-N-alkylation of the neomycin B outmaneuvers the aminoglycoside resistant enzyme acetyltransferase(3)IIIa via an unexpected mechanism
To Be Published
|
|
