2VEC
 
 | | The crystal structure of the protein YhaK from Escherichia coli | | Descriptor: | CHLORIDE ION, PIRIN-LIKE PROTEIN YHAK | | Authors: | Gurmu, D, Lu, J, Johnson, K.A, Nordlund, P, Holmgren, A, Erlandsen, H. | | Deposit date: | 2007-10-18 | | Release date: | 2008-07-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of the Protein Yhak from Escherichia Coli Reveals a New Subclass of Redox Sensitive Enterobacterial Bicupins.
Proteins, 74, 2008
|
|
3FHD
 
 | | Crystal structure of the Shutoff and Exonuclease Protein from Kaposis Sarcoma Associated Herpesvirus | | Descriptor: | MAGNESIUM ION, ORF 37, SULFATE ION | | Authors: | Dahlroth, S.L, Gurmu, D, Schmitzberger, F, Haas, J, Erlandsen, H, Nordlund, P. | | Deposit date: | 2008-12-09 | | Release date: | 2009-11-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the shutoff and exonuclease protein from the oncogenic Kaposi's sarcoma-associated herpesvirus
Febs J., 276, 2009
|
|
2G6V
 
 | | The crystal structure of ribD from Escherichia coli | | Descriptor: | Riboflavin biosynthesis protein ribD | | Authors: | Stenmark, P, Moche, M, Gurmu, D, Nordlund, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-02-25 | | Release date: | 2007-02-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of the Bifunctional Deaminase/Reductase RibD of the Riboflavin Biosynthetic Pathway in Escherichia coli: Implications for the Reductive Mechanism.
J.Mol.Biol., 373, 2007
|
|
1XA4
 
 | | Crystal structure of CaiB, a type III CoA transferase in carnitine metabolism | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COENZYME A, Crotonobetainyl-CoA:carnitine CoA-transferase, ... | | Authors: | Stenmark, P, Gurmu, D, Nordlund, P. | | Deposit date: | 2004-08-25 | | Release date: | 2004-11-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of CaiB, a Type-III CoA Transferase in Carnitine Metabolism
Biochemistry, 43, 2004
|
|
1XA3
 
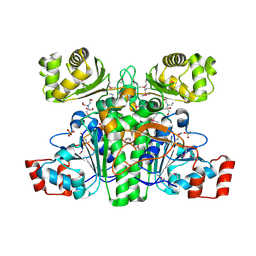 | | Crystal structure of CaiB, a type III CoA transferase in carnitine metabolism | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Crotonobetainyl-CoA:carnitine CoA-transferase, SULFATE ION | | Authors: | Stenmark, P, Gurmu, D, Nordlund, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-08-25 | | Release date: | 2004-11-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of CaiB, a Type-III CoA Transferase in Carnitine Metabolism
Biochemistry, 43, 2004
|
|
2OBC
 
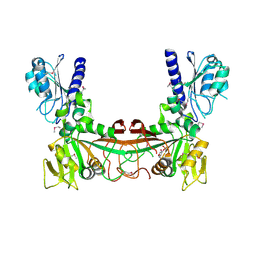 | | The crystal structure of RibD from Escherichia coli in complex with a substrate analogue, ribose 5-phosphate (beta form), bound to the active site of the reductase domain | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, Riboflavin biosynthesis protein ribD | | Authors: | Moche, M, Stenmark, P, Gurmu, D, Nordlund, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-12-18 | | Release date: | 2007-02-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the bifunctional deaminase/reductase RibD of the riboflavin biosynthetic pathway in Escherichia coli: implications for the reductive mechanism.
J.Mol.Biol., 373, 2007
|
|
2O7P
 
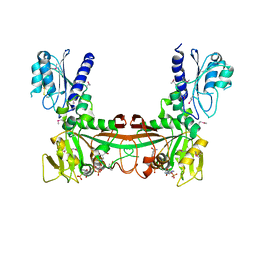 | | The crystal structure of RibD from Escherichia coli in complex with the oxidised NADP+ cofactor in the active site of the reductase domain | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Riboflavin biosynthesis protein ribD | | Authors: | Moche, M, Stenmark, P, Gurmu, D, Nordlund, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-12-11 | | Release date: | 2007-02-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the bifunctional deaminase/reductase RibD of the riboflavin biosynthetic pathway in Escherichia coli: implications for the reductive mechanism.
J.Mol.Biol., 373, 2007
|
|
