6X7Z
 
 | | Inositol-bound structure of Marinomonas primoryensis PA14 carbohydrate-binding domain | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1,2-ETHANEDIOL, Antifreeze protein, ... | | Authors: | Guo, S, Davies, P.L. | | Deposit date: | 2020-06-01 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural Basis of Ligand Selectivity by a Bacterial Adhesin Lectin Involved in Multispecies Biofilm Formation.
Mbio, 12, 2021
|
|
6X9P
 
 | |
6X8A
 
 | |
6X8D
 
 | |
6XA5
 
 | |
6X95
 
 | |
6X9M
 
 | |
6X7Y
 
 | |
6X8Y
 
 | |
6XAC
 
 | |
6X7X
 
 | |
6XAQ
 
 | |
6X7T
 
 | |
6X7J
 
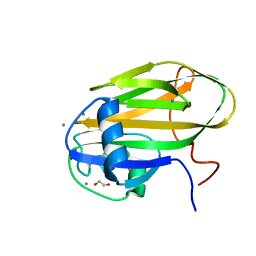 | |
5JUH
 
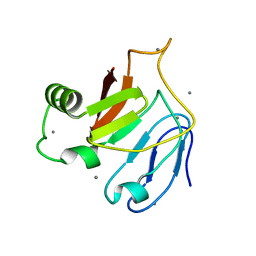 | | Crystal structure of C-terminal domain (RV) of MpAFP | | Descriptor: | Antifreeze protein, CALCIUM ION | | Authors: | Guo, S. | | Deposit date: | 2016-05-10 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
5K8G
 
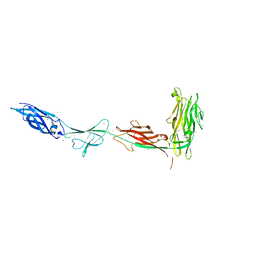 | |
3EB7
 
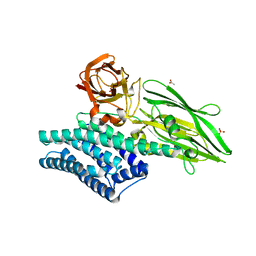 | | Crystal Structure of Insecticidal Delta-Endotoxin Cry8Ea1 from Bacillus Thuringiensis at 2.2 Angstroms Resolution | | Descriptor: | ACETATE ION, Insecticidal Delta-Endotoxin Cry8Ea1, SULFATE ION | | Authors: | Guo, S, Ye, S, Song, F, Zhang, J, Wei, L, Shu, C.L. | | Deposit date: | 2008-08-27 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Bacillus thuringiensis Cry8Ea1: An insecticidal toxin toxic to underground pests, the larvae of Holotrichia parallela.
J.Struct.Biol., 168, 2009
|
|
5IRB
 
 | | Structural insight into host cell surface retention of a 1.5-MDa bacterial ice-binding adhesin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Guo, S, Phippen, S, Campbell, R, Davies, P. | | Deposit date: | 2016-03-12 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
4KDW
 
 | | Crystal structure of a bacterial immunoglobulin-like domain from the M. primoryensis ice-binding adhesin | | Descriptor: | Antifreeze protein, CALCIUM ION, GLYCEROL | | Authors: | Guo, S, Garnham, C.P, Karunan, S.P, Campbell, R.L, Allingham, J.S, Davies, P.L. | | Deposit date: | 2013-04-25 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Role of Ca(2+) in folding the tandem beta-sandwich extender domains of a bacterial ice-binding adhesin.
Febs J., 280, 2013
|
|
4KDV
 
 | | Crystal structure of a bacterial immunoglobulin-like domain from the M. primoryensis ice-binding adhesin | | Descriptor: | Antifreeze protein, CALCIUM ION | | Authors: | Guo, S, Garnham, C.P, Karunan, S.P, Campbell, R.L, Allingham, J.S, Davies, P.L. | | Deposit date: | 2013-04-25 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Role of Ca(2+) in folding the tandem beta-sandwich extender domains of a bacterial ice-binding adhesin.
Febs J., 280, 2013
|
|
5IX9
 
 | | Cell surface anchoring domain | | Descriptor: | Antifreeze protein | | Authors: | Guo, S, Langelaan, D. | | Deposit date: | 2016-03-23 | | Release date: | 2017-06-28 | | Last modified: | 2020-01-08 | | Method: | SOLUTION NMR | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
5J6Y
 
 | | Crystal structure of PA14 domain of MpAFP Antifreeze protein | | Descriptor: | Antifreeze protein, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Guo, S. | | Deposit date: | 2016-04-05 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
3CX9
 
 | | Crystal Structure of Human serum albumin complexed with Myristic acid and lysophosphatidylethanolamine | | Descriptor: | (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, MYRISTIC ACID, Serum albumin | | Authors: | Guo, S, Yang, F, Chen, L, Bian, C, Huang, M. | | Deposit date: | 2008-04-24 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of transport of lysophospholipids by human serum albumin.
Biochem.J., 423, 2009
|
|
6X6M
 
 | |
6X5V
 
 | |
