4R9I
 
 | | Crystal structure of cysteine proteinase inhibitor Serpin18 from Bombyx mori | | Descriptor: | BETA-MERCAPTOETHANOL, CITRATE ANION, SODIUM ION, ... | | Authors: | Guo, P.C, He, H.W, Zhao, P, Xia, Q.Y. | | Deposit date: | 2014-09-05 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into the unique inhibitory mechanism of the silkworm protease inhibitor serpin18
Sci Rep, 5, 2015
|
|
8IUI
 
 | |
4E0H
 
 | | Crystal structure of FAD binding domain of Erv1 from Saccharomyces cerevisiae | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Mitochondrial FAD-linked sulfhydryl oxidase ERV1 | | Authors: | Guo, P.C, Ma, J.D, Jiang, Y.L, Wang, S.J, Hu, T.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2012-03-04 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of yeast sulfhydryl oxidase erv1 reveals electron transfer of the disulfide relay system in the mitochondrial intermembrane space
J.Biol.Chem., 287, 2012
|
|
4E0I
 
 | | Crystal structure of the C30S/C133S mutant of Erv1 from Saccharomyces cerevisiae | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Mitochondrial FAD-linked sulfhydryl oxidase ERV1 | | Authors: | Guo, P.C, Ma, J.D, Jiang, Y.L, Wang, S.J, Hu, T.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2012-03-04 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of yeast sulfhydryl oxidase erv1 reveals electron transfer of the disulfide relay system in the mitochondrial intermembrane space
J.Biol.Chem., 287, 2012
|
|
4PVC
 
 | |
4PVD
 
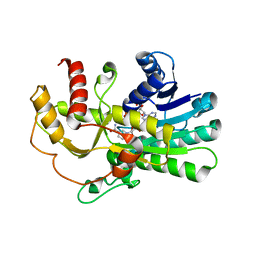 | | Crystal structure of yeast methylglyoxal/isovaleraldehyde reductase Gre2 complexed with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent methylglyoxal reductase GRE2 | | Authors: | Guo, P.C, Bao, Z.Z, Li, W.F, Zhou, C.Z. | | Deposit date: | 2014-03-17 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the cofactor-assisted substrate recognition of yeast methylglyoxal/isovaleraldehyde reductase Gre2
Biochim.Biophys.Acta, 1844, 2014
|
|
3QWB
 
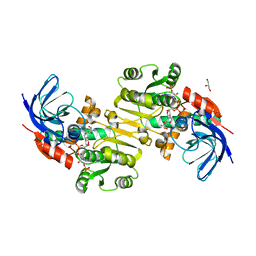 | | Crystal structure of Saccharomyces cerevisiae Zeta-crystallin-like quinone oxidoreductase Zta1 complexed with NADPH | | Descriptor: | GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Probable quinone oxidoreductase | | Authors: | Guo, P.C, Ma, X.X, Bao, Z.Z, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2011-02-28 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural insights into the cofactor-assisted substrate recognition of yeast quinone oxidoreductase Zta1
J.Struct.Biol., 176, 2011
|
|
3QWA
 
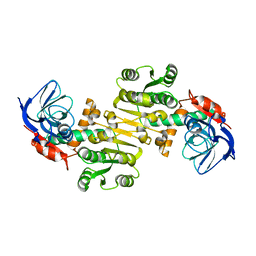 | | Crystal structure of Saccharomyces cerevisiae Zeta-crystallin-like quinone oxidoreductase Zta1 | | Descriptor: | Probable quinone oxidoreductase | | Authors: | Guo, P.C, Ma, X.X, Bao, Z.Z, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2011-02-27 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the cofactor-assisted substrate recognition of yeast quinone oxidoreductase Zta1
J.Struct.Biol., 176, 2011
|
|
7WNS
 
 | |
7EJB
 
 | |
4IJC
 
 | | Crystal structure of arabinose dehydrogenase Ara1 from Saccharomyces cerevisiae | | Descriptor: | D-arabinose dehydrogenase [NAD(P)+] heavy chain, GLYCEROL, SULFATE ION | | Authors: | Hu, X.Q, Guo, P.C, Li, W.F, Zhou, C.Z. | | Deposit date: | 2012-12-21 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Saccharomyces cerevisiaeD-arabinose dehydrogenase Ara1 and its complex with NADPH: implications for cofactor-assisted substrate recognition
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4IJR
 
 | | Crystal structure of Saccharomyces cerevisiae arabinose dehydrogenase Ara1 complexed with NADPH | | Descriptor: | D-arabinose dehydrogenase [NAD(P)+] heavy chain, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hu, X.Q, Guo, P.C, Li, W.F, Zhou, C.Z. | | Deposit date: | 2012-12-23 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Saccharomyces cerevisiaeD-arabinose dehydrogenase Ara1 and its complex with NADPH: implications for cofactor-assisted substrate recognition
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3PIL
 
 | | Crystal structure of Mxr1 from Saccharomyces cerevisiae in reduced form | | Descriptor: | ACETATE ION, Peptide methionine sulfoxide reductase | | Authors: | Ma, X.X, Guo, P.C, Shi, W.W, Luo, M, Tan, X.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2010-11-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural plasticity of the thioredoxin recognition site of yeast methionine S-sulfoxide reductase Mxr1
J.Biol.Chem., 286, 2011
|
|
3PIN
 
 | | Crystal structure of Mxr1 from Saccharomyces cerevisiae in complex with Trx2 | | Descriptor: | Peptide methionine sulfoxide reductase, Thioredoxin-2 | | Authors: | Ma, X.X, Guo, P.C, Shi, W.W, Luo, M, Tan, X.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2010-11-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural plasticity of the thioredoxin recognition site of yeast methionine S-sulfoxide reductase Mxr1
J.Biol.Chem., 286, 2011
|
|
3PIM
 
 | | Crystal structure of Mxr1 from Saccharomyces cerevisiae in unusual oxidized form | | Descriptor: | Peptide methionine sulfoxide reductase | | Authors: | Ma, X.X, Guo, P.C, Shi, W.W, Luo, M, Tan, X.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2010-11-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural plasticity of the thioredoxin recognition site of yeast methionine S-sulfoxide reductase Mxr1
J.Biol.Chem., 286, 2011
|
|
7V2S
 
 | | Crystal structure of juvenile hormone acid methyltransferase JHAMT isoform3 from silkworm | | Descriptor: | Methyltranfer_dom domain-containing protein | | Authors: | Guo, P.C, Zhang, Y.S, Zhang, l, Xu, H.Y. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | Structural characterization and functional analysis of juvenile hormone acid methyltransferase JHAMT3 from the silkworm, Bombyx mori.
Insect Biochem.Mol.Biol., 151, 2022
|
|
7VEO
 
 | | Crystal structure of juvenile hormone acid methyltransferase silkworm JHAMT isoform3 complex with S-Adenosyl-L-homocysteine | | Descriptor: | Methyltranfer_dom domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Guo, P.C, Zhang, Y.S, Zhang, l, Xu, H.Y. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural characterization and functional analysis of juvenile hormone acid methyltransferase JHAMT3 from the silkworm, Bombyx mori.
Insect Biochem.Mol.Biol., 151, 2022
|
|
7XND
 
 | | Crystal structure of Phosphomevalonate kinase from Silkworm | | Descriptor: | GLYCEROL, Phosphomevalonate kinase | | Authors: | Guo, P.C, Zhang, H. | | Deposit date: | 2022-04-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the substrate binding of phosphomevalonate kinase from the silkworm, Bombyx mori.
Insect Biochem.Mol.Biol., 150, 2022
|
|
7EBX
 
 | | Crystal structure of juvenile hormone acid methyltransferase JHAMT in complex with S-adenosyl-L-homocysteine. | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, juvenile hormone acid methyltransferase | | Authors: | Guo, P.C, Zhang, Y.S, Zhang, L, Xu, H.Y, Xia, Q.Y. | | Deposit date: | 2021-03-11 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural basis for juvenile hormone biosynthesis by the juvenile hormone acid methyltransferase.
J.Biol.Chem., 297, 2021
|
|
7EC0
 
 | | Crystal structure of juvenile hormone acid methyltransferase JHAMT in complex with S-Adenosyl homocysteine and methyl farnesoate | | Descriptor: | Juvenile hormone acid methyltransferase, Methyl farnesoate, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Guo, P.C, Zhang, Y.S, Zhang, L, Xu, H.Y, Xia, Q.Y. | | Deposit date: | 2021-03-11 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Structural basis for juvenile hormone biosynthesis by the juvenile hormone acid methyltransferase.
J.Biol.Chem., 297, 2021
|
|
7EBS
 
 | | Crystal structure of juvenile hormone acid methyltransferase JHAMT from silkworm | | Descriptor: | Juvenile hormone acid methyltransferase | | Authors: | Guo, P.C, Zhang, Y.S, Zhang, L, Xu, H.Y, Xia, Q.Y. | | Deposit date: | 2021-03-11 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for juvenile hormone biosynthesis by the juvenile hormone acid methyltransferase.
J.Biol.Chem., 297, 2021
|
|
3QUW
 
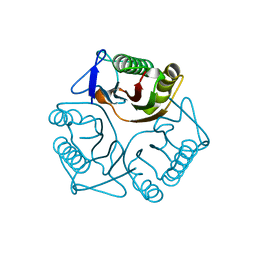 | | Crystal structure of yeast Mmf1 | | Descriptor: | Protein MMF1 | | Authors: | Jiang, Y.L, Pu, Y.G, Ma, X.X, Chen, Y, Zhou, C.Z. | | Deposit date: | 2011-02-24 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures and putative interface of Saccharomyces cerevisiae mitochondrial matrix proteins Mmf1 and Mam33.
J.Struct.Biol., 175, 2011
|
|
3QV0
 
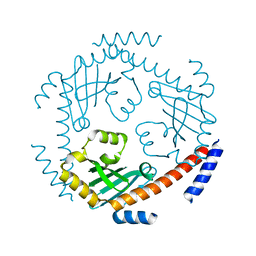 | | Crystal structure of Saccharomyces cerevisiae Mam33 | | Descriptor: | Mitochondrial acidic protein MAM33 | | Authors: | Jiang, Y.L, Pu, Y.G, Ma, X.X, Chen, Y, Zhou, C.Z. | | Deposit date: | 2011-02-24 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures and putative interface of Saccharomyces cerevisiae mitochondrial matrix proteins Mmf1 and Mam33.
J.Struct.Biol., 175, 2011
|
|
6KTH
 
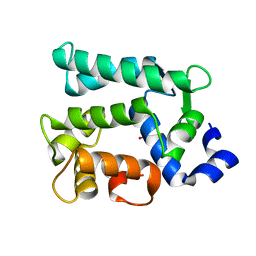 | | Crystal structure of Juvenile hormone diol kinase JHDK-L2 from silkworm, Bombyx mori | | Descriptor: | CALCIUM ION, GLYCEROL, Juvenile hormone diol kinase | | Authors: | Zhang, Y.S, Xu, H.Y, Wang, Z, Zhang, L, Zhao, P, Guo, P.C. | | Deposit date: | 2019-08-28 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structural characterization and functional analysis of juvenile hormone diol kinase from the silkworm, Bombyx mori.
Int.J.Biol.Macromol., 167, 2021
|
|
