7PJD
 
 | |
5OV5
 
 | | Bacillus megaterium porphobilinogen deaminase D82E mutant | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Guo, J, Erskine, P, Coker, A.R, Wood, S.P, Cooper, J.B. | | Deposit date: | 2017-08-28 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural studies of domain movement in active-site mutants of porphobilinogen deaminase from Bacillus megaterium.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5OT1
 
 | | The type III pullulan hydrolase from Thermococcus kodakarensis | | Descriptor: | CALCIUM ION, Pullulanase type II, GH13 family | | Authors: | Guo, J, Coker, A.R, Wood, S.P, Cooper, J.B, Keegan, R, Ahmad, N, Muhammad, M.A, Rashid, N, Akhtar, M. | | Deposit date: | 2017-08-18 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and function of the type III pullulan hydrolase from Thermococcus kodakarensis.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5OV6
 
 | | Bacillus megaterium porphobilinogen deaminase D82N mutant | | Descriptor: | 3-[4-(2-hydroxy-2-oxoethyl)-2,5-dimethyl-1~{H}-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | Authors: | Guo, J, Erskine, P, Coker, A.R, Wood, S.P, Cooper, J.B. | | Deposit date: | 2017-08-28 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural studies of domain movement in active-site mutants of porphobilinogen deaminase from Bacillus megaterium.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5OT0
 
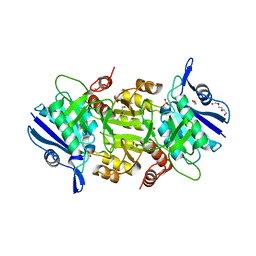 | | The thermostable L-asparaginase from Thermococcus kodakarensis | | Descriptor: | 1,2-ETHANEDIOL, L-asparaginase, PHOSPHATE ION, ... | | Authors: | Guo, J, Coker, A.R, Wood, S.P, Cooper, J.B, Rashid, N, Chohan, S.M, Akhtar, M. | | Deposit date: | 2017-08-18 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure and function of the thermostable L-asparaginase from Thermococcus kodakarensis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5OV4
 
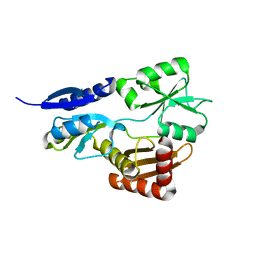 | | Bacillus megaterium porphobilinogen deaminase D82A mutant | | Descriptor: | Porphobilinogen deaminase | | Authors: | Guo, J, Erskine, P, Coker, A.R, Wood, S.P, Cooper, J.B. | | Deposit date: | 2017-08-28 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Structural studies of domain movement in active-site mutants of porphobilinogen deaminase from Bacillus megaterium.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
6P2I
 
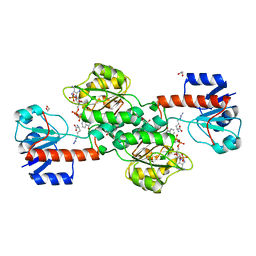 | | Acyclic imino acid reductase (Bsp5) in complex with NADPH and D-Arg | | Descriptor: | 1,2-ETHANEDIOL, D-ARGININE, Glycerate dehydrogenase, ... | | Authors: | Guo, J, Higgins, M.A, Daniel-Ivad, P, Ryan, K.S. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | An Asymmetric Reductase That Intercepts Acyclic Imino Acids Producedin Situby a Partner Oxidase.
J.Am.Chem.Soc., 141, 2019
|
|
7POJ
 
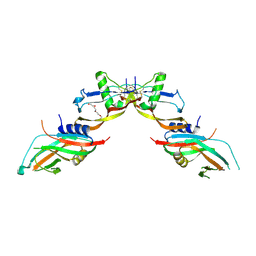 | | Prodomain bound BMP10 crystal form 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bone morphogenetic protein 10, TETRAETHYLENE GLYCOL | | Authors: | Guo, J, Yu, M, Li, W. | | Deposit date: | 2021-09-09 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of BMPRII extracellular domain in binary and ternary receptor complexes with BMP10.
Nat Commun, 13, 2022
|
|
7POI
 
 | | Prodomain bound BMP10 crystal form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bone morphogenetic protein 10, D(-)-TARTARIC ACID | | Authors: | Guo, J, Yu, M, Li, W. | | Deposit date: | 2021-09-09 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of BMPRII extracellular domain in binary and ternary receptor complexes with BMP10.
Nat Commun, 13, 2022
|
|
7PPC
 
 | | Ternary signalling complex of BMP10 bound to ALK1 and BMPRII | | Descriptor: | Bone morphogenetic protein 10, Bone morphogenetic protein receptor type-2, Serine/threonine-protein kinase receptor R3 | | Authors: | Guo, J, Yu, M, Read, R.J, Li, W. | | Deposit date: | 2021-09-13 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structures of BMPRII extracellular domain in binary and ternary receptor complexes with BMP10.
Nat Commun, 13, 2022
|
|
7PPB
 
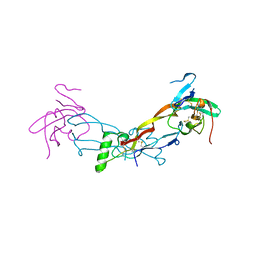 | |
7PPA
 
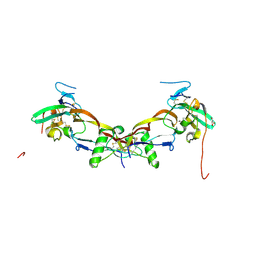 | | High resolution structure of bone morphogenetic protein receptor type II (BMPRII) extracellular domain in complex with BMP10 | | Descriptor: | Bone morphogenetic protein 10, Bone morphogenetic protein receptor type-2, GLYCEROL | | Authors: | Guo, J, Yu, M, Read, R.J, Li, W. | | Deposit date: | 2021-09-13 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structures of BMPRII extracellular domain in binary and ternary receptor complexes with BMP10.
Nat Commun, 13, 2022
|
|
5MDN
 
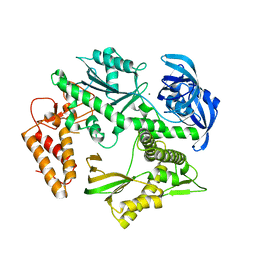 | | Structure of the family B DNA polymerase from the hyperthermophilic archaeon Pyrobaculum calidifontis | | Descriptor: | DNA polymerase, MAGNESIUM ION | | Authors: | Guo, J, Zhang, W, Coker, A.R, Wood, S.P, Cooper, J.B, Rashid, N, Akhtar, M. | | Deposit date: | 2016-11-12 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the family B DNA polymerase from the hyperthermophilic archaeon Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6T6W
 
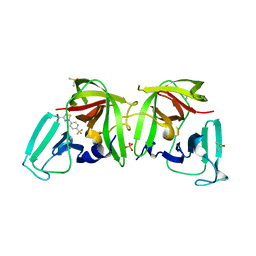 | |
6TCF
 
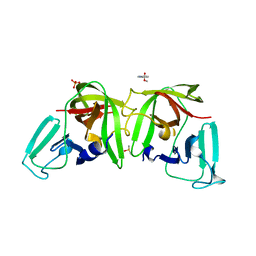 | |
6T82
 
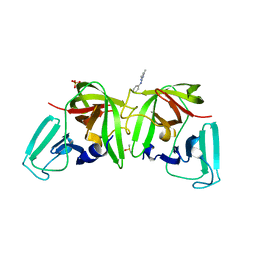 | |
6T8T
 
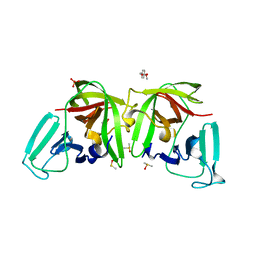 | |
6TC1
 
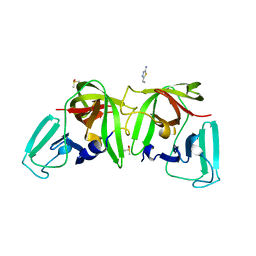 | |
6TGL
 
 | |
6T1Q
 
 | | 3C-like protease from Southampton norovirus. | | Descriptor: | Genome polyprotein | | Authors: | Guo, J, Cooper, J.B. | | Deposit date: | 2019-10-05 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | In crystallo-screening for discovery of human norovirus 3C-like protease inhibitors.
J Struct Biol X, 4, 2020
|
|
6T2I
 
 | |
6T4E
 
 | |
6T5R
 
 | |
6T8R
 
 | | 3C-like protease from Southampton virus complexed with FMOPL000605a. | | Descriptor: | 4-(5-amino-1,3,4-thiadiazol-2-yl)phenol, DIMETHYL SULFOXIDE, Genome polyprotein, ... | | Authors: | Guo, J, Cooper, J.B. | | Deposit date: | 2019-10-24 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | In crystallo-screening for discovery of human norovirus 3C-like protease inhibitors.
J Struct Biol X, 4, 2020
|
|
6TBO
 
 | |
