1HSB
 
 | | DIFFERENT LENGTH PEPTIDES BIND TO HLA-AW68 SIMILARLY AT THEIR ENDS BUT BULGE OUT IN THE MIDDLE | | Descriptor: | ALANINE, ARGININE, BETA 2-MICROGLOBULIN, ... | | Authors: | Guo, H.-C, Strominger, J.L, Wiley, D.C. | | Deposit date: | 1993-03-30 | | Release date: | 1993-10-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Different length peptides bind to HLA-Aw68 similarly at their ends but bulge out in the middle.
Nature, 360, 1992
|
|
1YFI
 
 | | Crystal Structure of restriction endonuclease MspI in complex with its cognate DNA in P212121 space group | | Descriptor: | 5'-D(*CP*CP*CP*CP*CP*GP*GP*GP*GP*G)-3', Type II restriction enzyme MspI | | Authors: | Xu, Q.S, Kucera, R.B, Roberts, R.J, Guo, H.-C. | | Deposit date: | 2005-01-02 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two crystal forms of the restriction enzyme MspI-DNA complex show the same novel structure.
Protein Sci., 14, 2005
|
|
5J5E
 
 | | crystal structure of antigen-ERAP1 domain complex | | Descriptor: | Endoplasmic reticulum aminopeptidase 1 | | Authors: | Sui, L, Gandhi, A, Guo, H.-C. | | Deposit date: | 2016-04-02 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a polypeptide's C-terminus in complex with the regulatory domain of ER aminopeptidase 1.
Mol.Immunol., 80, 2016
|
|
2GAW
 
 | |
2GAC
 
 | |
9GAF
 
 | |
9GAC
 
 | |
9GAA
 
 | |
3RJO
 
 | |
3LJQ
 
 | |
1TMC
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF A CLASS I MAJOR HISTOCOMPATIBILITY COMPLEX MOLECULE MISSING THE ALPHA3 DOMAIN OF THE HEAVY CHAIN | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-AW68), DECAMERIC PEPTIDE (EVAPPEYHRK) | | Authors: | Collins, E.J, Garboczi, D.N, Karpusas, M.N, Wiley, D.C. | | Deposit date: | 1994-12-19 | | Release date: | 1995-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three-dimensional structure of a class I major histocompatibility complex molecule missing the alpha 3 domain of the heavy chain.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
2ZAK
 
 | | Orthorhombic crystal structure of precursor E. coli isoaspartyl peptidase/L-asparaginase (EcAIII) with active-site T179A mutation | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, L-asparaginase precursor, ... | | Authors: | Michalska, K, Hernandez-Santoyo, A, Jaskolski, M. | | Deposit date: | 2007-10-07 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal packing of plant-type L-asparaginase from Escherichia coli
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2ZAL
 
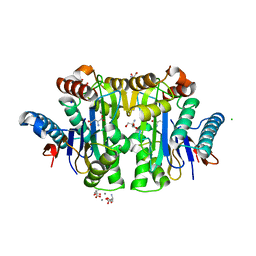 | | Crystal structure of E. coli isoaspartyl aminopeptidase/L-asparaginase in complex with L-aspartate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ASPARTIC ACID, CALCIUM ION, ... | | Authors: | Michalska, K, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2007-10-07 | | Release date: | 2007-10-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of isoaspartyl aminopeptidase in complex with L-aspartate
J.Biol.Chem., 280, 2005
|
|
3C17
 
 | |
2CLR
 
 | |
