1ICF
 
 | | CRYSTAL STRUCTURE OF MHC CLASS II ASSOCIATED P41 II FRAGMENT IN COMPLEX WITH CATHEPSIN L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (CATHEPSIN L: HEAVY CHAIN), PROTEIN (CATHEPSIN L: LIGHT CHAIN), ... | | Authors: | Guncar, G, Pungercic, G, Klemencic, I, Turk, V, Turk, D. | | Deposit date: | 1999-01-07 | | Release date: | 2000-01-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of MHC class II-associated p41 Ii fragment bound to cathepsin L reveals the structural basis for differentiation between cathepsins L and S.
EMBO J., 18, 1999
|
|
8PCH
 
 | | CRYSTAL STRUCTURE OF PORCINE CATHEPSIN H DETERMINED AT 2.1 ANGSTROM RESOLUTION: LOCATION OF THE MINI-CHAIN C-TERMINAL CARBOXYL GROUP DEFINES CATHEPSIN H AMINOPEPTIDASE FUNCTION | | Descriptor: | CATHEPSIN H, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Guncar, G, Podobnik, M, Pungercar, J, Strukelj, B, Turk, V, Turk, D. | | Deposit date: | 1997-11-07 | | Release date: | 1998-12-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of porcine cathepsin H determined at 2.1 A resolution: location of the mini-chain C-terminal carboxyl group defines cathepsin H aminopeptidase function.
Structure, 6, 1998
|
|
1EF7
 
 | | CRYSTAL STRUCTURE OF HUMAN CATHEPSIN X | | Descriptor: | CATHEPSIN X | | Authors: | Guncar, G, Klemencic, I, Turk, B, Turk, V, Karaoglanovic-Carmona, A, Juliano, L, Turk, D. | | Deposit date: | 2000-02-07 | | Release date: | 2000-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of cathepsin X: a flip-flop of the ring of His23 allows carboxy-monopeptidase and carboxy-dipeptidase activity of the protease.
Structure Fold.Des., 8, 2000
|
|
2OPC
 
 | | Structure of Melampsora lini avirulence protein, AvrL567-A | | Descriptor: | AvrL567-A, COBALT (II) ION, IMIDAZOLE | | Authors: | Guncar, G, Wang, C.I, Forwood, J.K, Teh, T, Catanzariti, A.M, Ellis, J.G, Dodds, P.N, Kobe, B. | | Deposit date: | 2007-01-29 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The use of Co2+ for crystallization and structure determination, using a conventional monochromatic X-ray source, of flax rust avirulence protein.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2QVT
 
 | | Structure of Melampsora lini avirulence protein, AvrL567-D | | Descriptor: | AvrL567-D | | Authors: | Guncar, G, Wang, C.I, Forwood, J.K, Teh, T, Catanzariti, A.M, Lawrence, G, Schirra, H.J, Anderson, P.A, Ellis, J.G, Dodds, P.N, Kobe, B. | | Deposit date: | 2007-08-08 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of flax rust avirulence proteins AvrL567-A and -D reveal details of the structural basis for flax disease resistance specificity.
Plant Cell, 19, 2007
|
|
2Q2B
 
 | |
1TZQ
 
 | | Crystal structure of the equinatoxin II 8-69 double cysteine mutant | | Descriptor: | Equinatoxin II | | Authors: | Kristan, K, Podlesek, Z, Hojnik, V, Gutirrez-Aguirre, I, Guncar, G, Turk, D.A, Gonzalez-Maas, J.M, Lakey, J.H, Anderluh, G. | | Deposit date: | 2004-07-11 | | Release date: | 2004-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pore formation by equinatoxin, a eukaryotic pore-forming toxin, requires a flexible N-terminal region and a stable beta-sandwich
J.Biol.Chem., 279, 2004
|
|
2V1O
 
 | | Crystal structure of N-terminal domain of acyl-CoA thioesterase 7 | | Descriptor: | COENZYME A, CYTOSOLIC ACYL COENZYME A THIOESTER HYDROLASE | | Authors: | Forwood, J.K, Thakur, A.S, Guncar, G, Marfori, M, Mouradov, D, Meng, W.N, Robinson, J, Huber, T, Kellie, S, Martin, J.L, Hume, D.A, Kobe, B. | | Deposit date: | 2007-05-28 | | Release date: | 2007-06-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for Recruitment of Tandem Hotdog Domains in Acyl-Coa Thioesterase 7 and its Role in Inflammation.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
3EA5
 
 | | Kap95p Binding Induces the Switch Loops of RanGDP to adopt the GTP-bound Conformation: Implications for Nuclear Import Complex Assembly Dynamics | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, Importin subunit beta-1, ... | | Authors: | Forwood, J.K, Lonhienne, J.K, Guncar, G, Stewart, M, Marfori, M, Kobe, B. | | Deposit date: | 2008-08-24 | | Release date: | 2008-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kap95p binding induces the switch loops of RanGDP to adopt the GTP-bound conformation: implications for nuclear import complex assembly dynamics.
J.Mol.Biol., 383, 2008
|
|
4MZV
 
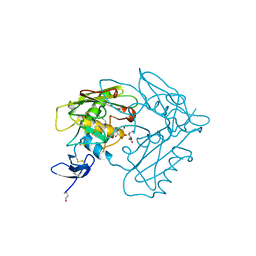 | | Crystal structure of extracellular part of human EpCAM | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, Epithelial cell adhesion molecule | | Authors: | Pavsic, M, Guncar, G, Djinovic-Carugo, K, Lenarcic, B. | | Deposit date: | 2013-09-30 | | Release date: | 2014-08-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.865 Å) | | Cite: | Crystal structure and its bearing towards an understanding of key biological functions of EpCAM.
Nat Commun, 5, 2014
|
|
3L9S
 
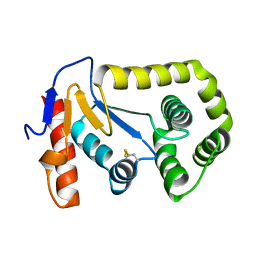 | |
3DIH
 
 | |
3L9U
 
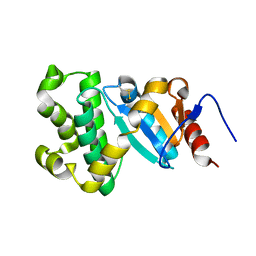 | |
3L9V
 
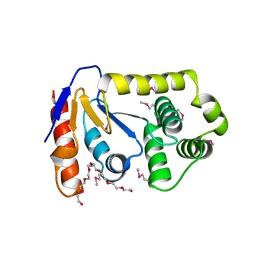 | | Crystal Structure of Salmonella enterica serovar Typhimurium SrgA | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, HEXAETHYLENE GLYCOL, O-ACETALDEHYDYL-HEXAETHYLENE GLYCOL, ... | | Authors: | Heras, B, Jarrott, R, Shouldice, S.R, Guncar, G. | | Deposit date: | 2010-01-05 | | Release date: | 2010-03-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Structural and functional characterization of three DsbA paralogues from Salmonella enterica serovar typhimurium
J.Biol.Chem., 285, 2010
|
|
2OCT
 
 | |
1NB5
 
 | | Crystal structure of stefin A in complex with cathepsin H | | Descriptor: | Cathepsin H, Cathepsin H MINI CHAIN, STEFIN A, ... | | Authors: | Jenko, S, Dolenc, I, Guncar, G, Dobersek, A, Podobnik, M, Turk, D. | | Deposit date: | 2002-12-02 | | Release date: | 2003-02-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of stefin A in complex with cathepsin H: N-terminal residues of inhibitors can adapt to the active sites of endo- and exopeptidases
J.Mol.Biol., 326, 2003
|
|
1NB3
 
 | | Crystal structure of stefin A in complex with cathepsin H: N-terminal residues of inhibitors can adapt to the active sites of endo-and exopeptidases | | Descriptor: | CATHEPSIN H MINI CHAIN, Cathepsin H, Stefin A, ... | | Authors: | Jenko, S, Dolenc, I, Guncar, G, Dobersek, A, Podobnik, M, Turk, D. | | Deposit date: | 2002-12-02 | | Release date: | 2003-02-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of stefin A in complex with cathepsin H: N-terminal residues of inhibitors can adapt to the active sites of endo- and exopeptidases
J.Mol.Biol., 326, 2003
|
|
