7SG3
 
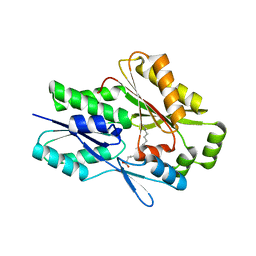 | | The X-ray crystal structure of the Staphylococcus aureus Fatty Acid Kinase B1 mutant A121I-A158L to 2.35 Angstrom resolution (Open form chain A, Palmitate bound) | | Descriptor: | Fatty Acid Kinase B1, PALMITIC ACID | | Authors: | Cuypers, M.G, Gullett, J.M, Rock, C.O, White, S.W. | | Deposit date: | 2021-10-04 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of structural transitions in bacterial fatty acid binding proteins that permit ligand entry and exit at membranes.
J.Biol.Chem., 298, 2022
|
|
7SCL
 
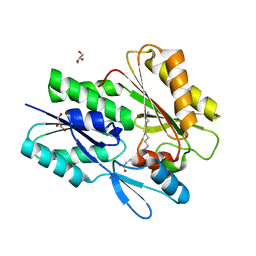 | | The X-ray crystal structure of Staphylococcus aureus Fatty Acid Kinase B1 (FakB1) mutant R173A in complex with Palmitate to 1.60 Angstrom resolution | | Descriptor: | Fatty Acid Kinase B1, GLYCEROL, PALMITIC ACID | | Authors: | Cuypers, M.G, Gullett, J.M, Subramanian, C, Rock, C.O, White, S.W. | | Deposit date: | 2021-09-28 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of structural transitions in bacterial fatty acid binding proteins that permit ligand entry and exit at membranes.
J.Biol.Chem., 298, 2022
|
|
6NM1
 
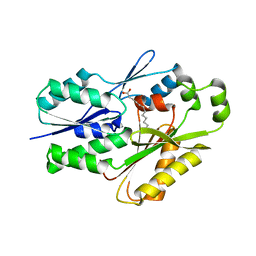 | | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein A158L mutant to 2.33 Angstrom resolution exhibits a conformation change compared to the wild type form | | Descriptor: | Fatty acid Kinase (Fak) B1 protein, MYRISTIC ACID | | Authors: | Cuypers, M.G, Gullett, J.M, Subramanian, C, Ericson, M, White, S.W, Rock, C.O. | | Deposit date: | 2019-01-10 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Identification of structural transitions in bacterial fatty acid binding proteins that permit ligand entry and exit at membranes.
J.Biol.Chem., 298, 2022
|
|
6NOK
 
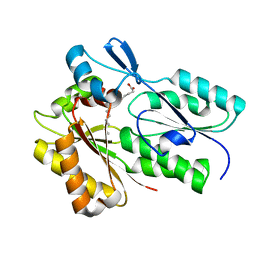 | | The X-ray crystal structure of Streptococcus pneumoniae Fatty Acid Kinase (Fak) B1 protein loaded with myristic acid (C14:0) to 1.69 Angstrom resolution | | Descriptor: | Fatty Acid Kinase (Fak) B1 protein, MYRISTIC ACID, SODIUM ION | | Authors: | Cuypers, M.G, Gullett, J.M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2019-01-16 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The X-ray crystal structure of Streptococcus pneumoniae Fatty Acid Kinase (Fak) B1 protein loaded with myristic acid (C14:0) to 1.69 Angstrom resolution
To Be Published
|
|
6NYU
 
 | | The X-ray crystal structure of Staphylococcus aureus Fatty Acid Kinase (Fak) B1 F263T mutant protein to 2.18 Angstrom resolution | | Descriptor: | DegV domain-containing protein, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Cuypers, M.G, Gullett, J.M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2019-02-12 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.183 Å) | | Cite: | The X-ray crystal structure of STAPHYLOCOCCUS AUREUS Fatty Acid Kinase (Fak) B1 protein F263T mutant to 2.18 Angstrom resolution
To Be Published
|
|
6NR1
 
 | | The X-ray crystal structure of Streptococcus pneumoniae Fatty Acid Kinase (Fak) B2 protein loaded with Vaccenic acid (C18:1 delta11) to 2.1 Angstrom resolution | | Descriptor: | Fatty Acid Kinase (Fak) B2 protein (SPR1019), GLYCEROL, VACCENIC ACID | | Authors: | Cuypers, M.G, Gullett, J.M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2019-01-22 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The X-ray crystal structure of Streptococcus pneumoniae Fatty Acid Kinase (Fak) B2 protein loaded with Vaccenic acid (C18:1 delta11) to 2.1 Angstrom resolution
To Be Published
|
|
6MH9
 
 | | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein A121I mutant to 2.02 Angstrom resolution | | Descriptor: | Fatty Acid Kinase (Fak) B1 protein, PALMITIC ACID | | Authors: | Cuypers, M.G, Ericson, M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2018-09-17 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Identification of structural transitions in bacterial fatty acid binding proteins that permit ligand entry and exit at membranes.
J.Biol.Chem., 298, 2022
|
|
5WOO
 
 | | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with Myristic acid (C14:0) to 1.78 Angstrom resolution | | Descriptor: | EDD domain protein, DegV family, GLYCEROL, ... | | Authors: | Cuypers, M.G, Ericson, M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2017-08-02 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Acyl-chain selectivity and physiological roles ofStaphylococcus aureusfatty acid-binding proteins.
J. Biol. Chem., 294, 2019
|
|
6ALW
 
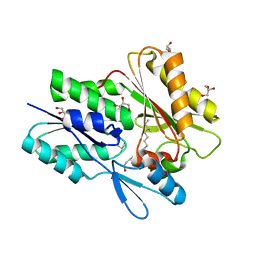 | | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with 12-Methyl Myristic Acid (C15:0) to 1.63 Angstrom resolution | | Descriptor: | (12R)-12-methyltetradecanoic acid, (12S)-12-methyltetradecanoic acid, EDD domain protein, ... | | Authors: | Cuypers, M.G, Ericson, M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2017-08-08 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Acyl-chain selectivity and physiological roles ofStaphylococcus aureusfatty acid-binding proteins.
J. Biol. Chem., 294, 2019
|
|
6B9I
 
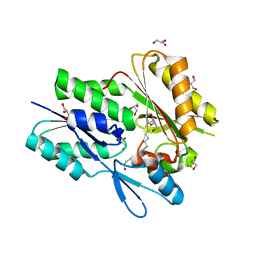 | | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with 14-Methylhexadecanoic Acid (Anteiso C17:0) to 1.93 Angstrom resolution | | Descriptor: | (14S)-14-methylhexadecanoic acid, Fatty acid Kinase (Fak) B1, GLYCEROL, ... | | Authors: | Cuypers, M.G, Ericson, M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2017-10-10 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Acyl-chain selectivity and physiological roles ofStaphylococcus aureusfatty acid-binding proteins.
J. Biol. Chem., 294, 2019
|
|
