1T5R
 
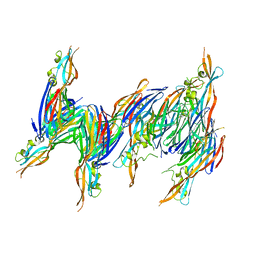 | | STRUCTURE OF THE PANTON-VALENTINE LEUCOCIDIN S COMPONENT FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | LukS-PV | | Authors: | Guillet, V, Roblin, P, Keller, D, Prevost, G, Mourey, L. | | Deposit date: | 2004-05-05 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of leucotoxin S component: new insight into the Staphylococcal beta-barrel pore-forming toxins.
J.Biol.Chem., 279, 2004
|
|
5MTW
 
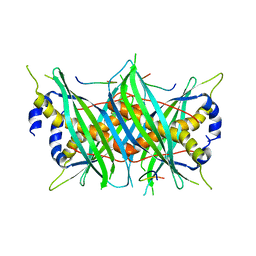 | |
1BGS
 
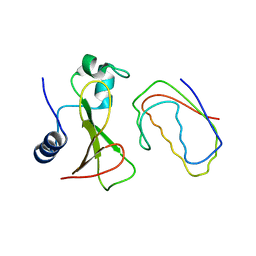 | | RECOGNITION BETWEEN A BACTERIAL RIBONUCLEASE, BARNASE, AND ITS NATURAL INHIBITOR, BARSTAR | | Descriptor: | BARNASE, BARSTAR | | Authors: | Guillet, V, Lapthorn, A, Mauguen, Y. | | Deposit date: | 1993-11-02 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition between a bacterial ribonuclease, barnase, and its natural inhibitor, barstar.
Structure, 1, 1993
|
|
5EY9
 
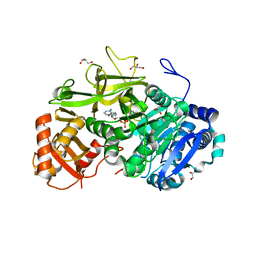 | | Structure of FadD32 from Mycobacterium marinum complexed to AMPC12 | | Descriptor: | GLYCEROL, Long-chain-fatty-acid--AMP ligase FadD32, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl icosyl hydrogen phosphate | | Authors: | Guillet, V, Maveyraud, L, Mourey, L. | | Deposit date: | 2015-11-24 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into Structure-Function Relationships and Inhibition of the Fatty Acyl-AMP Ligase (FadD32) Orthologs from Mycobacteria.
J.Biol.Chem., 291, 2016
|
|
5EY8
 
 | | Structure of FadD32 from Mycobacterium smegmatis complexed to AMPC20 | | Descriptor: | Acyl-CoA synthase, GLYCEROL, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl icosyl hydrogen phosphate | | Authors: | Guillet, V, Maveyraud, L, Mourey, L. | | Deposit date: | 2015-11-24 | | Release date: | 2015-12-16 | | Last modified: | 2016-04-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Insight into Structure-Function Relationships and Inhibition of the Fatty Acyl-AMP Ligase (FadD32) Orthologs from Mycobacteria.
J.Biol.Chem., 291, 2016
|
|
1MAV
 
 | | CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR DIVK AT PH 6.0 IN COMPLEX WITH MN2+ | | Descriptor: | MANGANESE (II) ION, cell division response regulator DivK | | Authors: | Guillet, V, Ohta, N, Cabantous, S, Newton, A, Samama, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-08-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic and Biochemical Studies of DivK Reveal Novel Features of
an Essential Response Regulator in Caulobacter crescentus.
J.Biol.Chem., 277, 2002
|
|
1MB3
 
 | | CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR DIVK AT PH 8.5 IN COMPLEX WITH MG2+ | | Descriptor: | MAGNESIUM ION, cell division response regulator DivK | | Authors: | Guillet, V, Ohta, N, Cabantous, S, Newton, A, Samama, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-08-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystallographic and Biochemical Studies of DivK Reveal Novel Features of
an Essential Response Regulator in Caulobacter crescentus.
J.Biol.Chem., 277, 2002
|
|
1M5U
 
 | | CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR DIVK. STRUCTURE AT PH 8.0 IN THE APO-FORM | | Descriptor: | cell division response regulator DivK | | Authors: | Guillet, V, Ohta, N, Cabantous, S, Newton, A, Samama, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-07-10 | | Release date: | 2002-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystallographic and biochemical studies of DivK reveal novel features of an essential response regulator in Caulobacter crescentus
J.Biol.Chem., 277, 2002
|
|
1M5T
 
 | | CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR DIVK | | Descriptor: | cell division response regulator DivK | | Authors: | Guillet, V, Ohta, N, Cabantous, S, Newton, A, Samama, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-07-10 | | Release date: | 2002-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic and biochemical studies of DivK reveal novel features of an essential response regulator in Caulobacter crescentus
J.Biol.Chem., 277, 2002
|
|
1MB0
 
 | | CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR DIVK AT PH 8.0 IN COMPLEX WITH MN2+ | | Descriptor: | MANGANESE (II) ION, cell division response regulator DivK | | Authors: | Guillet, V, Ohta, N, Cabantous, S, Newton, A, Samama, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-08-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and Biochemical Studies of DivK Reveal Novel Features of
an Essential Response Regulator in Caulobacter crescentus.
J.Biol.Chem., 277, 2002
|
|
3RIP
 
 | | Crystal Structure of human gamma-tubulin complex protein 4 (GCP4) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, GLYCEROL, Gamma-tubulin complex component 4 | | Authors: | Gregory-Pauron, L, Guillet, V, Mourey, L. | | Deposit date: | 2011-04-14 | | Release date: | 2011-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of gamma-tubulin complex protein GCP4 provides insight into microtubule nucleation.
Nat.Struct.Mol.Biol., 18, 2011
|
|
7AWK
 
 | |
1DCM
 
 | | STRUCTURE OF UNPHOSPHORYLATED FIXJ-N WITH AN ATYPICAL CONFORMER (MONOMER A) | | Descriptor: | MAGNESIUM ION, TRANSCRIPTIONAL REGULATORY PROTEIN FIXJ | | Authors: | Gouet, P, Fabry, B, Guillet, V, Birck, C, Mourey, L, Kahn, D, Samama, J.P. | | Deposit date: | 1999-11-05 | | Release date: | 2000-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural transitions in the FixJ receiver domain.
Structure Fold.Des., 7, 1999
|
|
1DBW
 
 | | CRYSTAL STRUCTURE OF FIXJ-N | | Descriptor: | POLYETHYLENE GLYCOL (N=34), TRANSCRIPTIONAL REGULATORY PROTEIN FIXJ | | Authors: | Gouet, P, Fabry, B, Guillet, V, Birck, C, Mourey, L, Kahn, D, Samama, J.P. | | Deposit date: | 1999-11-03 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural transitions in the FixJ receiver domain.
Structure Fold.Des., 7, 1999
|
|
1DCK
 
 | | STRUCTURE OF UNPHOSPHORYLATED FIXJ-N COMPLEXED WITH MN2+ | | Descriptor: | MANGANESE (II) ION, POLYETHYLENE GLYCOL (N=34), TRANSCRIPTIONAL REGULATORY PROTEIN FIXJ | | Authors: | Gouet, P, Fabry, B, Guillet, V, Birck, C, Mourey, L, Kahn, D, Samama, J.P. | | Deposit date: | 1999-11-05 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural transitions in the FixJ receiver domain.
Structure Fold.Des., 7, 1999
|
|
2QK7
 
 | | A covalent S-F heterodimer of staphylococcal gamma-hemolysin | | Descriptor: | Gamma-hemolysin component A, Gamma-hemolysin component B | | Authors: | Roblin, P, Guillet, V, Maveyraud, L, Mourey, L. | | Deposit date: | 2007-07-10 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A covalent S-F heterodimer of leucotoxin reveals molecular plasticity of beta-barrel pore-forming toxins.
Proteins, 71, 2008
|
|
2FK8
 
 | |
2FK7
 
 | |
1DY6
 
 | | Structure of the imipenem-hydrolyzing beta-lactamase SME-1 | | Descriptor: | CARBAPENEM-HYDROLYSING BETA-LACTAMASE SME-1 | | Authors: | Sougakoff, W, L'Hermite, G, Billy, I, Guillet, V, Naas, T, Nordman, P, Jarlier, V, Delettre, J. | | Deposit date: | 2000-01-27 | | Release date: | 2001-01-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure of the Imipenem-Hydrolyzing Class a Beta-Lactamase Sme-1 from Serratia Marcescens.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2H26
 
 | | human CD1b in complex with endogenous phosphatidylcholine and spacer | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Garcia-Alles, L.F, Maveyraud, L, Vallina, A.T, Guillet, V, Mourey, L. | | Deposit date: | 2006-05-18 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Endogenous phosphatidylcholine and a long spacer ligand stabilize the lipid-binding groove of CD1b.
Embo J., 25, 2006
|
|
1I5N
 
 | | Crystal structure of the P1 domain of CheA from Salmonella typhimurium | | Descriptor: | CHEMOTAXIS PROTEIN CHEA, SULFATE ION | | Authors: | Mourey, L, Da Re, S, Pedelacq, J.-D, Tolstyk, T, Faurie, C, Guillet, V, Stock, J.B, Samama, J.-P. | | Deposit date: | 2001-02-28 | | Release date: | 2001-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of the CheA histidine phosphotransfer domain that mediates response regulator phosphorylation in bacterial chemotaxis
J.Biol.Chem., 276, 2001
|
|
1BT5
 
 | | CRYSTAL STRUCTURE OF THE IMIPENEM INHIBITED TEM-1 BETA-LACTAMASE FROM ESCHERICHIA COLI | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, PROTEIN (BETA-LACTAMASE), SULFATE ION | | Authors: | Maveyraud, L, Mourey, L, Pedelacq, J.D, Guillet, V, Kotra, L.K, Mobashery, S, Samama, J.P. | | Deposit date: | 1998-09-02 | | Release date: | 1999-09-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Clinical Longevity of Carbapenem Antibiotics in the Face of Challenge by the Common Class A Beta-Lactamases from Antibiotic-Resistant Bacteria
J.Am.Chem.Soc., 120, 1998
|
|
1E25
 
 | | The high resolution structure of PER-1 class A beta-lactamase | | Descriptor: | EXTENDED-SPECTRUM BETA-LACTAMASE PER-1, SULFATE ION | | Authors: | Tranier, S, Bouthors, A.T, Maveyraud, L, Guillet, V, Sougakoff, W, Samama, J.P. | | Deposit date: | 2000-05-17 | | Release date: | 2000-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The High Resolution Crystal Structure for Class a Beta-Lactamase Per-1 Reveals the Bases for its Increase in Breadth of Activity
J.Biol.Chem., 275, 2000
|
|
1FFS
 
 | | CHEY-BINDING DOMAIN OF CHEA IN COMPLEX WITH CHEY FROM CRYSTALS SOAKED IN ACETYL PHOSPHATE | | Descriptor: | CHEMOTAXIS PROTEIN CHEA, CHEMOTAXIS PROTEIN CHEY, MANGANESE (II) ION | | Authors: | Gouet, P, Chinardet, N, Welch, M, Guillet, V, Birck, C, Mourey, L, Samama, J.-P. | | Deposit date: | 2000-07-26 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Further insights into the mechanism of function of the response regulator CheY from crystallographic studies of the CheY--CheA(124--257) complex.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1FFG
 
 | | CHEY-BINDING DOMAIN OF CHEA IN COMPLEX WITH CHEY AT 2.1 A RESOLUTION | | Descriptor: | CHEMOTAXIS PROTEIN CHEA, CHEMOTAXIS PROTEIN CHEY, MANGANESE (II) ION | | Authors: | Gouet, P, Chinardet, N, Welch, M, Guillet, V, Birck, C, Mourey, L, Samama, J.-P. | | Deposit date: | 2000-07-25 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Further insights into the mechanism of function of the response regulator CheY from crystallographic studies of the CheY--CheA(124--257) complex.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
