1DSR
 
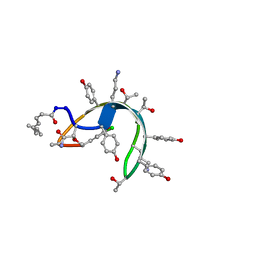 | | Peptide antibiotic, NMR, 6 structures | | Descriptor: | (2Z,4E)-7-methylocta-2,4-dienoic acid, RAMOPLANIN A2, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Kurz, M, Guba, W. | | Deposit date: | 1996-07-05 | | Release date: | 1997-02-12 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | 3D Structure of Ramoplanin: A Potent Inhibitor of Bacterial Cell Wall Synthesis.
Biochemistry, 35, 1996
|
|
6QZH
 
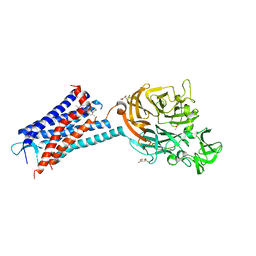 | | Structure of the human CC Chemokine Receptor 7 in complex with the intracellular allosteric antagonist Cmp2105 and the insertion protein Sialidase NanA | | Descriptor: | 3-[[4-[[(1~{R})-2,2-dimethyl-1-(5-methylfuran-2-yl)propyl]amino]-1,1-bis(oxidanylidene)-1,2,5-thiadiazol-3-yl]amino]-~{N},~{N},6-trimethyl-2-oxidanyl-benzamide, C-C chemokine receptor type 7,Sialidase A,C-C chemokine receptor type 7, D(-)-TARTARIC ACID, ... | | Authors: | Jaeger, K, Bruenle, S, Weinert, T, Guba, W, Muehle, J, Miyazaki, T, Weber, M, Furrer, A, Haenggi, N, Tetaz, T, Huang, C.Y, Mattle, D, Vonach, J.M, Gast, A, Kuglstatter, A, Rudolph, M.G, Nogly, P, Benz, J, Dawson, R.J.P, Standfuss, J. | | Deposit date: | 2019-03-11 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Allosteric Ligand Recognition in the Human CC Chemokine Receptor 7.
Cell, 178, 2019
|
|
6ZBV
 
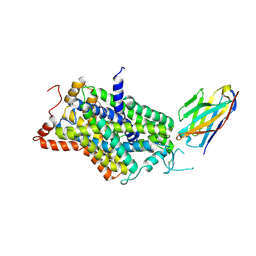 | | Inward-open structure of human glycine transporter 1 in complex with a benzoylisoindoline inhibitor and sybody Sb_GlyT1#7 | | Descriptor: | Sodium- and chloride-dependent glycine transporter 1,Sodium- and chloride-dependent glycine transporter 1, Sybody Sb_GlyT1#7, [5-fluoranyl-6-(oxan-4-yloxy)-1,3-dihydroisoindol-2-yl]-[5-methylsulfonyl-2-[2,2,3,3,3-pentakis(fluoranyl)propoxy]phenyl]methanone | | Authors: | Shahsavar, A, Stohler, P, Bourenkov, G, Zimmermann, I, Siegrist, M, Guba, W, Pinard, E, Sinning, S, Seeger, M.A, Schneider, T.R, Dawson, R.J.P, Nissen, P. | | Deposit date: | 2020-06-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insights into the inhibition of glycine reuptake.
Nature, 591, 2021
|
|
6T13
 
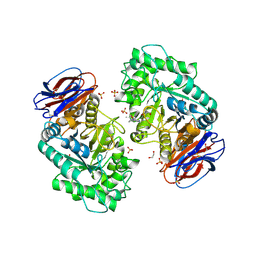 | | CRYSTAL STRUCTURE OF GLUCOCEREBROSIDASE IN COMPLEX WITH A PYRROLOPYRAZINE | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[2-(4-methoxyphenyl)-5-methyl-pyrrolo[2,3-b]pyrazin-6-yl]piperidin-1-yl]ethanone, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Benz, J, Ehler, A, Hug, M, Huber, S, Rufer, A.C, Guba, W, Jagasia, R, Hofmann, E.C, Rodriguez Sarmiento, R.M. | | Deposit date: | 2019-10-03 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Novel beta-Glucocerebrosidase Activators That Bind to a New Pocket at a Dimer Interface and Induce Dimerization.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6ZPL
 
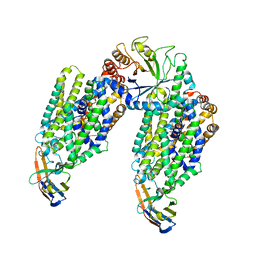 | | Inward-open structure of human glycine transporter 1 in complex with a benzoylisoindoline inhibitor, sybody Sb_GlyT1#7 and bound Na and Cl ions. | | Descriptor: | CHLORIDE ION, Endoglucanase H, SODIUM ION, ... | | Authors: | Shahsavar, A, Stohler, P, Bourenkov, G, Zimmermann, I, Siegrist, M, Guba, W, Pinard, E, Sinning, S, Seeger, M.A, Schneider, T.R, Dawson, R.J.P, Nissen, P. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.945 Å) | | Cite: | Structural insights into the inhibition of glycine reuptake.
Nature, 591, 2021
|
|
5I2R
 
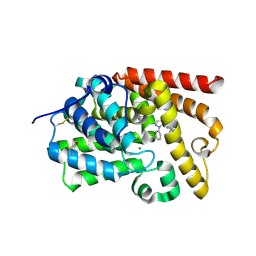 | | human PDE10A in complex with 3-(2-phenylpyrazol-3-yl)-1-[3-(trifluoromethoxy)phenyl]pyridazin-4-one | | Descriptor: | 3-(1-phenyl-1H-pyrazol-5-yl)-1-[3-(trifluoromethoxy)phenyl]pyridazin-4(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Koerner, M, Rudolph, M.G. | | Deposit date: | 2016-02-09 | | Release date: | 2016-03-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
6DMI
 
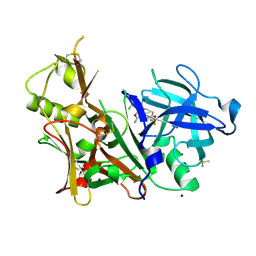 | | A multiconformer ligand model of 5T5 bound to BACE-1 | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, SODIUM ION, ... | | Authors: | Hudson, B.M, van Zundert, G, Keedy, D, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
3ZMG
 
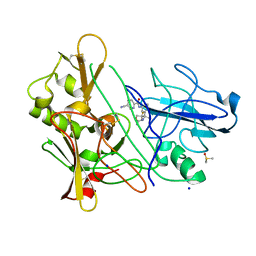 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH CHEMICAL LIGAND | | Descriptor: | BETA-SECRETASE 1, DIMETHYL SULFOXIDE, N-[3-[(4R)-2-azanylidene-5,5-bis(fluoranyl)-4-methyl-1,3-oxazinan-4-yl]-4-fluoranyl-phenyl]-5-cyano-pyridine-2-carboxamide, ... | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M. | | Deposit date: | 2013-02-08 | | Release date: | 2013-05-01 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Beta-Secretase (Bace1) Inhibitors with High in Vivo Efficacy Suitable for Clinical Evaluation in Alzheimer'S Disease.
J.Med.Chem., 56, 2013
|
|
3ZLQ
 
 | | BACE2 XAPERONE COMPLEX | | Descriptor: | 5-Ethoxy-pyridine-2-carboxylic acid [3-((R)-2-amino-5,5-difluoro-4-methyl-5,6-dihydro-4H-[1,3]oxazin-4-yl)-4-fluoro-phenyl]-amide, BETA-SECRETASE 2, XA4813 | | Authors: | Kuglstatter, A, Stihle, M. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-01 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Beta-Secretase (Bace1) Inhibitors with High in Vivo Efficacy Suitable for Clinical Evaluation in Alzheimer'S Disease.
J.Med.Chem., 56, 2013
|
|
4BFD
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH CHEMICAL LIGAND | | Descriptor: | BETA-SECRETASE 1, DIMETHYL SULFOXIDE, N-[3-[(1S,3S,6S)-5-azanyl-3-methyl-4-azabicyclo[4.1.0]hept-4-en-3-yl]-4-fluoranyl-phenyl]-5-chloranyl-pyridine-2-carbox amide, ... | | Authors: | Banner, D.W, Benz, J, Stihle, M. | | Deposit date: | 2013-03-18 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bace1 Inhibitors: A Head Group Scan on a Series of Amides.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4BEK
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH CHEMICAL LIGAND | | Descriptor: | (4S)-4-(4-methoxyphenyl)-4-methyl-5,6-dihydro-1,3-thiazin-2-amine, BETA-SECRETASE 1, DIMETHYL SULFOXIDE, ... | | Authors: | Banner, D.W, Benz, J, Stihle, M. | | Deposit date: | 2013-03-11 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Bace1 Inhibitors: A Head Group Scan on a Series of Amides.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5MCQ
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH ACTIVE SITE AND EXOSITE BINDING PEPTIDE INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, BACE-1 ACTIVE AND EXOSITE BINDING INHIBITOR, Beta-secretase 1 | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-09-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Potent and Selective BACE-1 Peptide Inhibitors Lower Brain A beta Levels Mediated by Brain Shuttle Transport.
EBioMedicine, 24, 2017
|
|
5MBW
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH Pep#3 | | Descriptor: | BACE1 INHIBITOR PEPTIDE Pep#3, Beta-secretase 1, CHLORIDE ION | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-09 | | Release date: | 2017-09-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Potent and Selective BACE-1 Peptide Inhibitors Lower Brain A beta Levels Mediated by Brain Shuttle Transport.
EBioMedicine, 24, 2017
|
|
5MCO
 
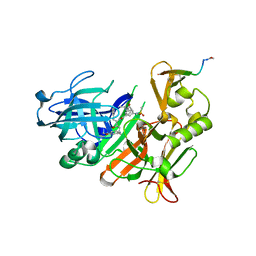 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH ACTIVE SITE INHIBITOR GRL-8234 AND EXOSITE PEPTIDE | | Descriptor: | BACE-1 EXOSITE PEPTIDE, Beta-secretase 1, N-{(1S,2R)-1-benzyl-2-hydroxy-3-[(3-methoxybenzyl)amino]propyl}-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-09-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Potent and Selective BACE-1 Peptide Inhibitors Lower Brain A beta Levels Mediated by Brain Shuttle Transport.
EBioMedicine, 24, 2017
|
|
5EDE
 
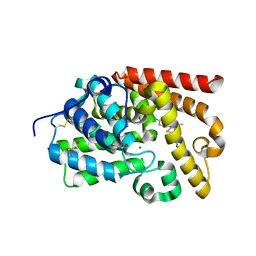 | | human PDE10A in complex with 1-(4-Chloro-phenyl)-3-methyl-1H-thieno[2,3-c]pyrazole-5-carboxylic acid (tetrahydro-furan-2-ylmethyl)-amide at 2.2A | | Descriptor: | 1-(4-chlorophenyl)-3-methyl-~{N}-[[(2~{R})-oxolan-2-yl]methyl]thieno[2,3-c]pyrazole-5-carboxamide, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Joseph, C, Rudolph, M.G. | | Deposit date: | 2015-10-21 | | Release date: | 2016-03-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
5EDH
 
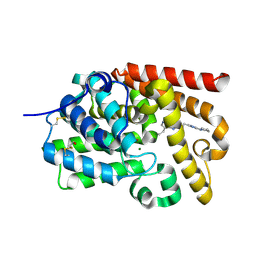 | | human PDE10A, 8-ethyl-5-methyl-2-[2-(2-methyl-5-pyrrolidin-1-yl-1,2,4-triazol-3-yl)ethyl]-[1,2,4]triazolo[1,5-c]pyrimidine, 2.03A, H3, Rfree=22.7% | | Descriptor: | 8-ethyl-5-methyl-2-[2-(2-methyl-5-pyrrolidin-1-yl-1,2,4-triazol-3-yl)ethyl]-[1,2,4]triazolo[1,5-c]pyrimidine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Rudolph, M.G. | | Deposit date: | 2015-10-21 | | Release date: | 2016-03-09 | | Last modified: | 2016-05-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
5EDI
 
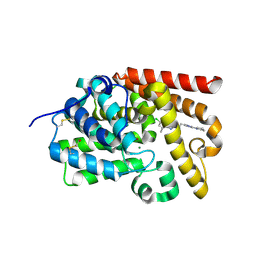 | | human PDE10A, 6-Chloro-5,8-dimethyl-2-[2-(2-methyl-5-pyrrolidin-1-yl-2H-[1,2,4]triazol-3-yl)-ethyl]-[1,2,4]triazolo[1,5-a]pyridine, 2.20A, H3, Rfree=23.5% | | Descriptor: | 6-chloranyl-5,8-dimethyl-2-[2-(2-methyl-5-pyrrolidin-1-yl-1,2,4-triazol-3-yl)ethyl]-[1,2,4]triazolo[1,5-a]pyridine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Rudolph, M.G. | | Deposit date: | 2015-10-21 | | Release date: | 2016-03-09 | | Last modified: | 2016-05-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
5EDC
 
 | |
5EDG
 
 | |
5EDB
 
 | |
5EZZ
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH (4S)-4-[3-(5-chloro-3-pyridyl)phenyl]-4-[4-(difluoromethoxy)-3-methyl-phenyl]-5H-oxazol-2-amine | | Descriptor: | (4~{S})-4-[4-[bis(fluoranyl)methoxy]-3-methyl-phenyl]-4-[3-(5-chloranylpyridin-3-yl)phenyl]-5~{H}-1,3-oxazol-2-amine, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Banner, D, Benz, J, Stihle, M, Kuglstatter, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-24 | | Last modified: | 2016-05-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
5F03
 
 | | TRYPTASE B2 IN COMPLEX WITH 5-(3-Aminomethyl-phenoxymethyl)-3-[3-(2-chloro-pyridin-3-ylethynyl)-phenyl]-oxazolidin-2-one; compound with trifluoro-acetic acid | | Descriptor: | (5~{S})-5-[[3-(aminomethyl)phenoxy]methyl]-3-[3-[2-(2-chloranylpyridin-3-yl)ethynyl]phenyl]-1,3-oxazolidin-2-one, Tryptase beta-2 | | Authors: | Banner, D, Benz, J, Joseph, C, Kuglstatter, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-24 | | Last modified: | 2019-03-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
5F00
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH 5-[3-[(3-chloro-8-quinolyl)amino]phenyl]-5-methyl-2,6-dihydro-1,4-oxazin-3-amine | | Descriptor: | (5~{R})-5-[3-[(3-chloranylquinolin-8-yl)amino]phenyl]-5-methyl-2,6-dihydro-1,4-oxazin-3-amine, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Banner, D, Benz, J, Stihle, M, Kuglstatter, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-24 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
5EZX
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH {(1R,2R)-2-[(R)-2-Amino-4-(4-difluoromethoxy-phenyl)-4,5-dihydro-oxazol-4-yl]-cyclopropyl}-(5-chloro-pyridin-3-yl)-methanone | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, SODIUM ION, ... | | Authors: | Banner, D, Benz, J, Stihle, M, Kuglstatter, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-24 | | Last modified: | 2016-05-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
5F02
 
 | | CATHEPSIN L IN COMPLEX WITH (2S,4R)-4-(2-Chloro-4-methoxy-benzenesulfonyl)-1-[3-(5-chloro-pyridin-2-yl)-azetidine-3-carbonyl]-pyrrolidine-2-carboxylic acid (1-cyano-cyclopropyl)-amide | | Descriptor: | (2~{S},4~{R})-4-[(2-chloranyl-4-methoxy-phenyl)-bis(oxidanyl)-$l^{4}-sulfanyl]-1-[3-(5-chloranylpyridin-2-yl)azetidin-3-yl]carbonyl-~{N}-[1-(iminomethyl)cyclopropyl]pyrrolidine-2-carboxamide, Cathepsin L1, GLYCEROL | | Authors: | Banner, D, Benz, J, Stihle, M, Kuglstatter, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-24 | | Last modified: | 2016-05-25 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
