1QOL
 
 | | STRUCTURE OF THE FMDV LEADER PROTEASE | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PROTEASE (NONSTRUCTURAL PROTEIN P20A) | | Authors: | Guarne, A, Tormo, J, Kirchweger, R, Pfistermueller, D, Skern, T, Fita, I. | | Deposit date: | 1999-11-13 | | Release date: | 2000-11-10 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Foot-and-Mouth Disease Virus Leader Protease: A Papain-Like Fold Adapted for Self-Processing and Eif4G Recognition.
Embo J., 17, 1998
|
|
1LRR
 
 | | CRYSTAL STRUCTURE OF E. COLI SEQA COMPLEXED WITH HEMIMETHYLATED DNA | | Descriptor: | 5'-D(*AP*GP*TP*CP*GP*(6MA)P*TP*CP*GP*GP*TP*G)-3', 5'-D(*CP*AP*CP*CP*GP*AP*TP*CP*GP*AP*CP*T)-3', SeqA protein | | Authors: | Guarne, A, Zhao, Q, Guirlando, R, Yang, W. | | Deposit date: | 2002-05-15 | | Release date: | 2002-12-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Insights into negative modulation of E. coli replication initiation from the structure of SeqA-hemimethylated DNA complex
NAT.STRUCT.BIOL., 9, 2002
|
|
1QMY
 
 | | FMDV LEADER PROTEASE (LBSHORT-C51A-C133S) | | Descriptor: | 1,2-ETHANEDIOL, PROTEASE | | Authors: | Guarne, A, Tormo, J, Glaser, W, Skern, T, Fita, I. | | Deposit date: | 1999-10-08 | | Release date: | 2000-10-12 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Biochemical Features Distinguish the Foot-and-Mouth Disease Virus Leader Proteinase from Other Papain-Like Enzymes
J.Mol.Biol., 302, 2000
|
|
1X9Z
 
 | | Crystal structure of the MutL C-terminal domain | | Descriptor: | CHLORIDE ION, DNA mismatch repair protein mutL, GLYCEROL, ... | | Authors: | Guarne, A, Ramon-Maiques, S, Wolff, E.M, Ghirlando, R, Hu, X, Miller, J.H, Yang, W. | | Deposit date: | 2004-08-24 | | Release date: | 2004-10-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the MutL C-terminal domain: a model of intact MutL and its roles in mismatch repair
Embo J., 23, 2004
|
|
1XRX
 
 | | Crystal structure of a DNA-binding protein | | Descriptor: | CALCIUM ION, SeqA protein | | Authors: | Guarne, A, Brendler, T, Zhao, Q, Ghirlando, R, Austin, S, Yang, W. | | Deposit date: | 2004-10-16 | | Release date: | 2005-05-10 | | Last modified: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of a SeqA-N filament: implications for DNA replication and chromosome organization.
Embo J., 24, 2005
|
|
1EA6
 
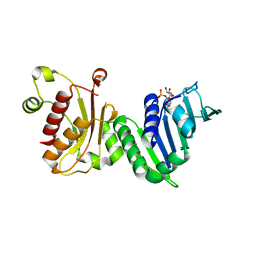 | |
1H7U
 
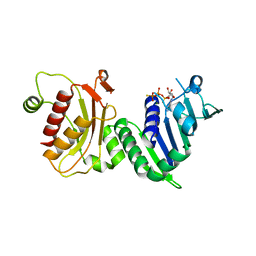 | | hPMS2-ATPgS | | Descriptor: | MAGNESIUM ION, MISMATCH REPAIR ENDONUCLEASE PMS2, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Guarne, A, Junop, M.S, Yang, W. | | Deposit date: | 2001-07-10 | | Release date: | 2001-11-27 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and Function of the N-Terminal 40 kDa Fragment of Human Pms2: A Monomeric Ghl ATPase
Embo J., 20, 2001
|
|
1H7S
 
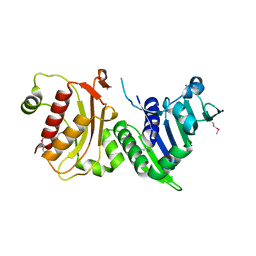 | | N-terminal 40kDa fragment of human PMS2 | | Descriptor: | PMS1 PROTEIN HOMOLOG 2 | | Authors: | Guarne, A, Junop, M.S, Yang, W. | | Deposit date: | 2001-07-10 | | Release date: | 2001-11-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Function of the N-Terminal 40 kDa Fragment of Human Pms2: A Monomeric Ghl ATPase
Embo J., 20, 2001
|
|
6BEK
 
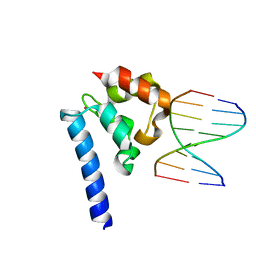 | |
4ITQ
 
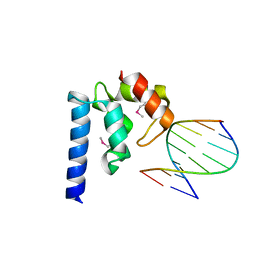 | | Crystal structure of hypothetical protein SCO1480 bound to DNA | | Descriptor: | 5'-D(P*CP*CP*GP*CP*GP*CP*GP*C)-3', 5'-D(P*GP*CP*GP*CP*GP*CP*GP*G)-3', Putative uncharacterized protein SCO1480 | | Authors: | Guarne, A, Nanji, T, Gloyd, M, Swiercz, J.P, Elliot, M.A. | | Deposit date: | 2013-01-18 | | Release date: | 2013-03-27 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A novel nucleoid-associated protein specific to the actinobacteria.
Nucleic Acids Res., 41, 2013
|
|
3U5V
 
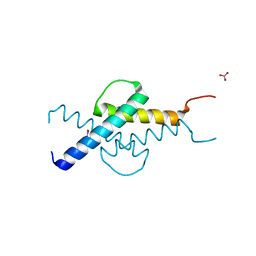 | | Crystal structure of Max-E47 | | Descriptor: | NITRATE ION, Protein max, Transcription factor E2-alpha chimera | | Authors: | Guarne, A, Ahmadpour, F, Gloyd, M. | | Deposit date: | 2011-10-11 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the minimalist max-e47 protein chimera.
Plos One, 7, 2012
|
|
6E8D
 
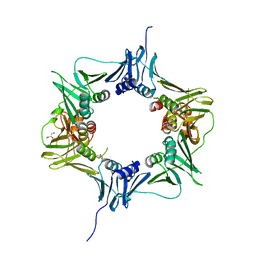 | |
6E8E
 
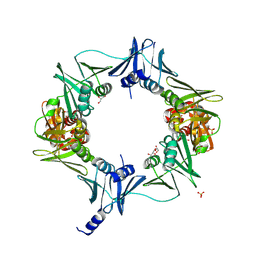 | |
3GAB
 
 | | C-terminal domain of Bacillus subtilis MutL crystal form I | | Descriptor: | DNA mismatch repair protein mutL | | Authors: | Guarne, A, Pillon, M.C, Lorenowicz, J.J, Mitchell, R.R, Chung, Y.S, Friedhoff, P. | | Deposit date: | 2009-02-17 | | Release date: | 2010-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the endonuclease domain of MutL: unlicensed to cut.
Mol.Cell, 39, 2010
|
|
5T2S
 
 | | Structure of the FHA1 domain of Rad53 bound simultaneously to the BRCT domain of Dbf4 and a phosphopeptide. | | Descriptor: | ASP-GLY-GLU-SER-TPO-ASP-GLU-ASP-ASP, DDK kinase regulatory subunit DBF4,Serine/threonine-protein kinase RAD53, GLYCEROL | | Authors: | Guarne, A, Almawi, A, Matthews, L. | | Deposit date: | 2016-08-24 | | Release date: | 2016-10-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 'AND' logic gates at work: Crystal structure of Rad53 bound to Dbf4 and Cdc7.
Sci Rep, 6, 2016
|
|
5D17
 
 | |
5T2F
 
 | | Structure of the FHA1 domain of Rad53 bound to the BRCT domain of Dbf4 | | Descriptor: | 1,2-ETHANEDIOL, DDK kinase regulatory subunit DBF4,Serine/threonine-protein kinase RAD53 chimeric protein | | Authors: | Guarne, A, Almawi, A.W, Matthews, L.A. | | Deposit date: | 2016-08-23 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | 'AND' logic gates at work: Crystal structure of Rad53 bound to Dbf4 and Cdc7.
Sci Rep, 6, 2016
|
|
3KDK
 
 | |
3KDG
 
 | |
5D16
 
 | |
3RPU
 
 | |
6MF5
 
 | |
6MF4
 
 | |
6MF6
 
 | |
6PPF
 
 | | Bacterial 45SRbgA ribosomal particle class B | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Ortega, J, Seffouh, A, Jain, N, Jahagirdar, D, Basu, K, Razi, A, Ni, X, Guarne, A, Britton, R.A. | | Deposit date: | 2019-07-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural consequences of the interaction of RbgA with a 50S ribosomal subunit assembly intermediate.
Nucleic Acids Res., 47, 2019
|
|
