2YN0
 
 | | tau55 histidine phosphatase domain | | Descriptor: | PHOSPHATE ION, TRANSCRIPTION FACTOR TAU 55 KDA SUBUNIT | | Authors: | Taylor, N.M.I, Glatt, S, Hennrich, M, von Scheven, G, Grotsch, H, Fernandez-Tornero, C, Rybin, V, Gavin, A.C, Kolb, P, Muller, C.W. | | Deposit date: | 2012-10-11 | | Release date: | 2013-04-03 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of a Phosphatase Domain within Yeast General Transcription Factor Iiic.
J.Biol.Chem., 288, 2013
|
|
2YN2
 
 | | Huf protein - paralogue of the tau55 histidine phosphatase domain | | Descriptor: | FORMIC ACID, UNCHARACTERIZED PROTEIN YNL108C | | Authors: | Taylor, N.M.I, Glatt, S, Hennrich, M, von Scheven, G, Grotsch, H, Fernandez-Tornero, C, Rybin, V, Gavin, A.C, Kolb, P, Muller, C.W. | | Deposit date: | 2012-10-11 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Functional Characterization of a Phosphatase Domain within Yeast General Transcription Factor Tfiiic.
J.Biol.Chem., 288, 2013
|
|
6HGC
 
 | | Structure of Calypso in complex with DEUBAD of ASX | | Descriptor: | Polycomb protein Asx, Ubiquitin carboxyl-terminal hydrolase calypso,Ubiquitin carboxyl-terminal hydrolase calypso | | Authors: | De, I, Chittock, E.C, Groetsch, H, Miller, T.C.R, McCarthy, A.A, Mueller, C.W. | | Deposit date: | 2018-08-23 | | Release date: | 2019-01-23 | | Last modified: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural Basis for the Activation of the Deubiquitinase Calypso by the Polycomb Protein ASX.
Structure, 27, 2019
|
|
7OB9
 
 | | Cryo-EM structure of human RNA Polymerase I in elongation state | | Descriptor: | DNA non-template strand, DNA template strand, DNA-directed RNA polymerase I subunit RPA1, ... | | Authors: | Misiaszek, A.D, Girbig, M, Mueller, C.W. | | Deposit date: | 2021-04-21 | | Release date: | 2021-12-08 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of human RNA polymerase I.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7OBB
 
 | | Cryo-EM structure of human RNA Polymerase I Open Complex | | Descriptor: | DNA non-template strand, DNA template strand, DNA-directed RNA polymerase I subunit RPA1, ... | | Authors: | Misiaszek, A.D, Girbig, M, Mueller, C.W. | | Deposit date: | 2021-04-21 | | Release date: | 2021-12-08 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of human RNA polymerase I.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7OBA
 
 | | Cryo-EM structure of human RNA Polymerase I in complex with RRN3 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA1, DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA2, ... | | Authors: | Misiaszek, A.D, Girbig, M, Mueller, C.W. | | Deposit date: | 2021-04-21 | | Release date: | 2021-12-08 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human RNA polymerase I.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6YJ6
 
 | | Structure of the TFIIIC subcomplex tauA | | Descriptor: | Transcription factor tau 131 kDa subunit, Transcription factor tau 55 kDa subunit, Transcription factor tau 95 kDa subunit,Transcription factor tau 95 kDa subunit | | Authors: | Vorlaender, M.K, Muller, C.W. | | Deposit date: | 2020-04-02 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the TFIIIC subcomplex tau A provides insights into RNA polymerase III pre-initiation complex formation.
Nat Commun, 11, 2020
|
|
7A6H
 
 | | Cryo-EM structure of human apo RNA Polymerase III | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Misiaszek, A.D, Vorlaender, M.K, Mueller, C.W. | | Deposit date: | 2020-08-25 | | Release date: | 2021-02-03 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AE1
 
 | | Cryo-EM structure of human RNA Polymerase III elongation complex 1 | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Misiaszek, A.D, Vorlaender, M.K, Mueller, C.W. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-03 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AE3
 
 | | Cryo-EM structure of human RNA Polymerase III elongation complex 3 | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Misiaszek, A.D, Vorlaender, M.K, Mueller, C.W. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-03 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AEA
 
 | | Cryo-EM structure of human RNA Polymerase III elongation complex 2 | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Misiaszek, A.D, Vorlaender, M.K, Mueller, C.W. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-03 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7Z1O
 
 | | Structure of yeast RNA Polymerase III PTC + NTPs | | Descriptor: | CHAPSO, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1N
 
 | | Structure of yeast RNA Polymerase III Delta C53-C37-C11 | | Descriptor: | CHAPSO, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1L
 
 | | Structure of yeast RNA Polymerase III Pre-Termination Complex (PTC) | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1M
 
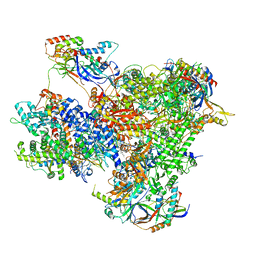 | | Structure of yeast RNA Polymerase III Elongation Complex (EC) | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
