1TRH
 
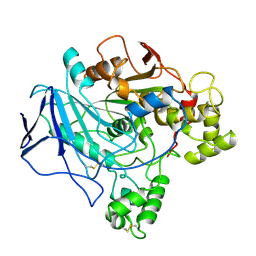 | |
1CIY
 
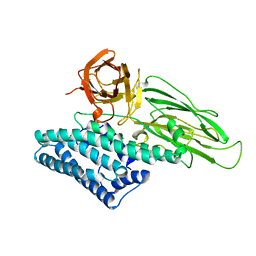 | |
1CRL
 
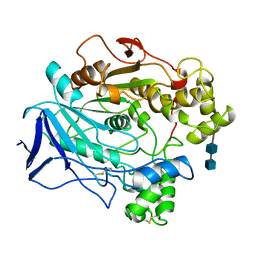 | |
3RAR
 
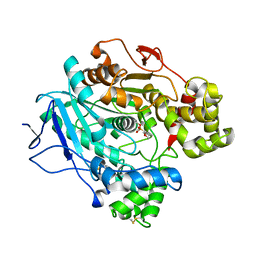 | |
1AJ5
 
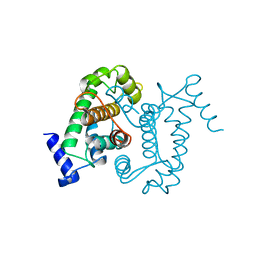 | | CALPAIN DOMAIN VI APO | | Descriptor: | CALPAIN | | Authors: | Cygler, M, Grochulski, P, Blanchard, H. | | Deposit date: | 1997-05-15 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a calpain Ca(2+)-binding domain reveals a novel EF-hand and Ca(2+)-induced conformational changes.
Nat.Struct.Biol., 4, 1997
|
|
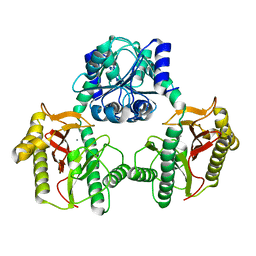
6XMR
 
 | |
4QPX
 
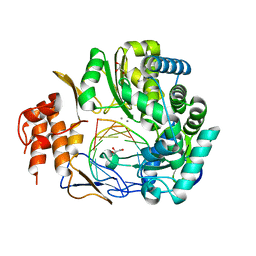 | | NV polymerase post-incorporation-like complex | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Polyprotein, ... | | Authors: | Zamyatkin, D.F, Parra, F, Grochulski, P, Ng, K.K.S. | | Deposit date: | 2014-06-25 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of a backtracked state reveals conformational changes similar to the state following nucleotide incorporation in human norovirus polymerase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
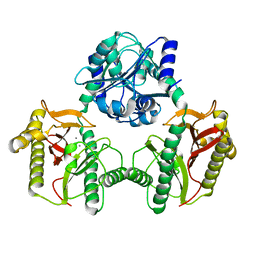
7N02
 
 | |
5V48
 
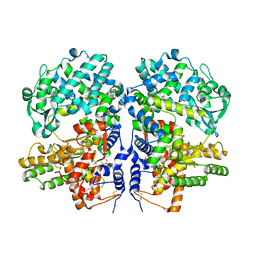 | | Soluble rabbit neprilysin in complex with thiorphan | | Descriptor: | (2-MERCAPTOMETHYL-3-PHENYL-PROPIONYL)-GLYCINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Labiuk, S.L, Grochulski, P, Sygusch, J. | | Deposit date: | 2017-03-08 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9965 Å) | | Cite: | Structures of soluble rabbit neprilysin complexed with phosphoramidon or thiorphan.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
4G54
 
 | |
7K3U
 
 | |
3H5X
 
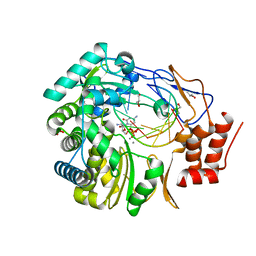 | | Crystal Structure of 2'-amino-2'-deoxy-cytidine-5'-triphosphate bound to Norovirus GII RNA polymerase | | Descriptor: | 2'-amino-2'-deoxycytidine 5'-(tetrahydrogen triphosphate), 5'-R(*UP*GP*CP*CP*CP*GP*GP*G)-3', 5'-R(P*UP*GP*CP*CP*CP*GP*GP*GP*C)-3', ... | | Authors: | Zamyatkin, D.F, Parra, F, Machin, A, Grochulski, P, Ng, K.K.S. | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Binding of 2'-amino-2'-deoxycytidine-5'-triphosphate to norovirus polymerase induces rearrangement of the active site.
J.Mol.Biol., 390, 2009
|
|
3H5Y
 
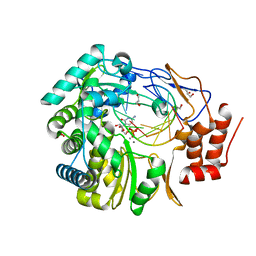 | | Norovirus polymerase+primer/template+CTP complex at 6 mM MnCl2 | | Descriptor: | 5'-R(*UP*GP*CP*CP*CP*GP*GP*G)-3', 5'-R(P*UP*GP*CP*CP*CP*GP*GP*GP*C)-3', CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Zamyatkin, D.F, Parra, F, Machin, A, Grochulski, P, Ng, K.K.S. | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Binding of 2'-amino-2'-deoxycytidine-5'-triphosphate to norovirus polymerase induces rearrangement of the active site.
J.Mol.Biol., 390, 2009
|
|
1NP8
 
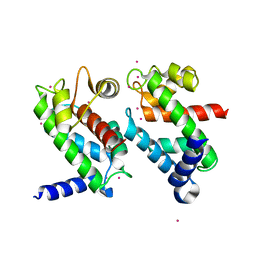 | | 18-k C-terminally trunucated small subunit of calpain | | Descriptor: | CADMIUM ION, Calcium-dependent protease, small subunit | | Authors: | Leinala, E.K, Arthur, J.S, Grochulski, P, Davies, P.L, Elce, J.S, Jia, Z. | | Deposit date: | 2003-01-17 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A second binding site revealed by C-terminal truncation of calpain small subunit, a penta-EF-hand protein
PROTEINS: STRUCT.,FUNCT.,GENET., 53, 2003
|
|
4XBH
 
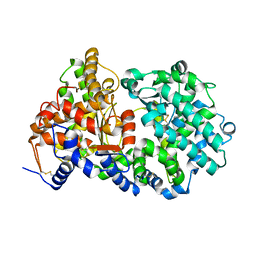 | | Soluble rabbit neprilysin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Neprilysin, ... | | Authors: | Labiuk, S.L, Grochulski, P, Sygusch, J. | | Deposit date: | 2014-12-16 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.114 Å) | | Cite: | Structures of soluble rabbit neprilysin complexed with phosphoramidon or thiorphan.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
4ZNG
 
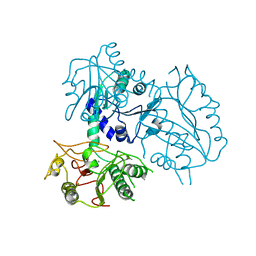 | |
4ZR5
 
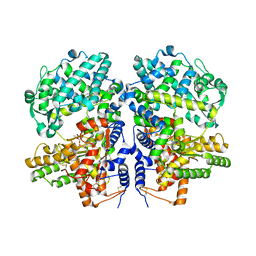 | | Soluble rabbit neprilysin in complex with phosphoramidon | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-ALPHA-L-RHAMNOPYRANOSYLOXY(HYDROXYPHOSPHINYL)-L-LEUCYL-L-TRYPTOPHAN, ... | | Authors: | Labiuk, S.L, Grochulski, P, Sygusch, J. | | Deposit date: | 2015-05-11 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8016 Å) | | Cite: | Structures of soluble rabbit neprilysin complexed with phosphoramidon or thiorphan.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
2I1Q
 
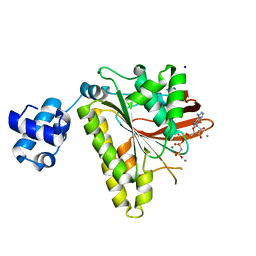 | | RadA Recombinase in complex with Calcium | | Descriptor: | CALCIUM ION, DNA repair and recombination protein radA, MAGNESIUM ION, ... | | Authors: | Qian, X, He, Y, Ma, X, Fodje, M.N, Grochulski, P, Luo, Y. | | Deposit date: | 2006-08-14 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Calcium Stiffens Archaeal Rad51 Recombinase from Methanococcus voltae for Homologous Recombination.
J.Biol.Chem., 281, 2006
|
|
1MIR
 
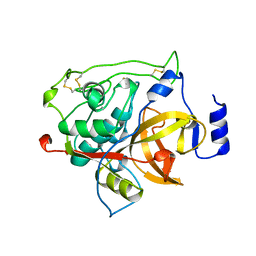 | | RAT PROCATHEPSIN B | | Descriptor: | PROCATHEPSIN B | | Authors: | Cygler, M, Sivaraman, J, Grochulski, P, Coulombe, R, Storer, A.C, Mort, J.S. | | Deposit date: | 1996-01-12 | | Release date: | 1997-01-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of rat procathepsin B: model for inhibition of cysteine protease activity by the proregion.
Structure, 4, 1996
|
|
1CJL
 
 | |
1DVI
 
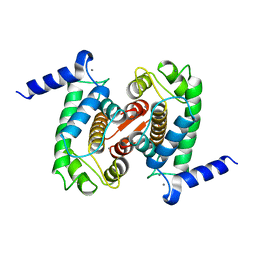 | | CALPAIN DOMAIN VI WITH CALCIUM BOUND | | Descriptor: | CALCIUM ION, CALPAIN | | Authors: | Cygler, M, Blanchard, H, Grochulski, P. | | Deposit date: | 1997-05-15 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a calpain Ca(2+)-binding domain reveals a novel EF-hand and Ca(2+)-induced conformational changes.
Nat.Struct.Biol., 4, 1997
|
|
1EUN
 
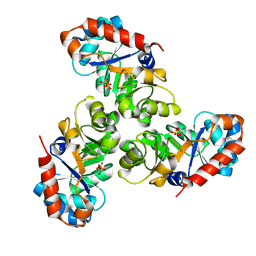 | |
1EUA
 
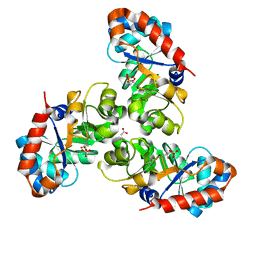 | | SCHIFF BASE INTERMEDIATE IN KDPG ALDOLASE FROM ESCHERICHIA COLI | | Descriptor: | ACETATE ION, KDPG ALDOLASE, PYRUVIC ACID, ... | | Authors: | Allard, J, Grochulski, P, Sygusch, J. | | Deposit date: | 2000-04-14 | | Release date: | 2001-02-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Covalent intermediate trapped in 2-keto-3-deoxy-6- phosphogluconate (KDPG) aldolase structure at 1.95-A resolution.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1F2E
 
 | | STRUCTURE OF SPHINGOMONAD, GLUTATHIONE S-TRANSFERASE COMPLEXED WITH GLUTATHIONE | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Nishio, T, Watanabe, T, Patel, A, Wang, Y, Lau, P.C.K, Grochulski, P, Li, Y, Cygler, M. | | Deposit date: | 2000-05-24 | | Release date: | 2000-06-21 | | Last modified: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Properties of a Sphingomonad and Marine Bacterium Beta-Class Glutathione S-Transferases and Crystal Structure of the Former Complex with Glutathione
To be published
|
|
1LPO
 
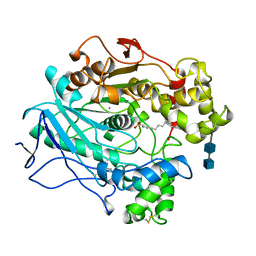 | |
