2BRD
 
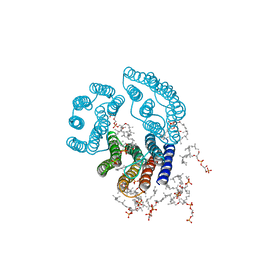 | | CRYSTAL STRUCTURE OF BACTERIORHODOPSIN IN PURPLE MEMBRANE | | Descriptor: | BACTERIORHODOPSIN, PHOSPHORIC ACID 2,3-BIS-(3,7,11,15-TETRAMETHYL-HEXADECYLOXY)-PROPYL ESTER 2-HYDROXO-3-PHOSPHONOOXY-PROPYL ESTER, RETINAL | | Authors: | Henderson, R, Grigorieff, N. | | Deposit date: | 1995-12-27 | | Release date: | 1996-06-10 | | Last modified: | 2012-05-30 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Electron-crystallographic refinement of the structure of bacteriorhodopsin.
J.Mol.Biol., 259, 1996
|
|
4V7Q
 
 | | Atomic model of an infectious rotavirus particle | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Core scaffold protein, ... | | Authors: | Settembre, E.C, Chen, J.Z, Dormitzer, P.R, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2010-05-13 | | Release date: | 2014-07-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic model of an infectious rotavirus particle.
Embo J., 30, 2011
|
|
4V7C
 
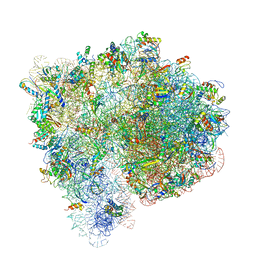 | | Structure of the Ribosome with Elongation Factor G Trapped in the Pre-Translocation State (pre-translocation 70S*tRNA structure) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Brilot, A.F, Korostelev, A.A, Ermolenko, D.N, Grigorieff, N. | | Deposit date: | 2013-11-20 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structure of the ribosome with elongation factor G trapped in the pretranslocation state.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4V7D
 
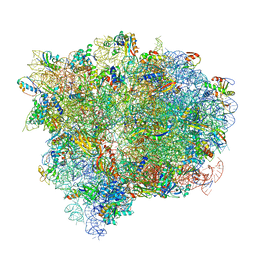 | | Structure of the Ribosome with Elongation Factor G Trapped in the Pre-Translocation State (pre-translocation 70S*tRNA*EF-G structure) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Brilot, A.F, Korostelev, A.A, Ermolenko, D.N, Grigorieff, N. | | Deposit date: | 2013-11-21 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structure of the ribosome with elongation factor G trapped in the pretranslocation state.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1XI5
 
 | | Clathrin D6 coat with auxilin J-domain | | Descriptor: | Auxilin J-domain, Clathrin heavy chain | | Authors: | Fotin, A, Cheng, Y, Grigorieff, N, Walz, T, Harrison, S.C, Kirchhausen, T. | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-02 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure of an auxilin-bound clathrin coat and its implications for the mechanism of uncoating
Nature, 432, 2004
|
|
1XI4
 
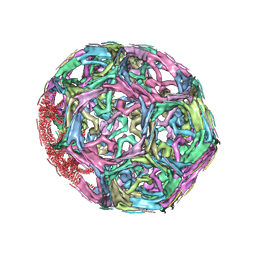 | | Clathrin D6 Coat | | Descriptor: | Clathrin heavy chain, Clathrin light chain A | | Authors: | Fotin, A, Cheng, Y, Sliz, P, Grigorieff, N, Harrison, S.C, Kirchhausen, T, Walz, T. | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-02 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Molecular model for a complete clathrin lattice from electron cryomicroscopy
Nature, 432, 2004
|
|
8DB3
 
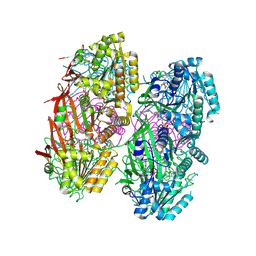 | | Crystal structure of KaiC with truncated C-terminal coiled-coil domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Circadian clock protein KaiC | | Authors: | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | Deposit date: | 2022-06-14 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
8DBA
 
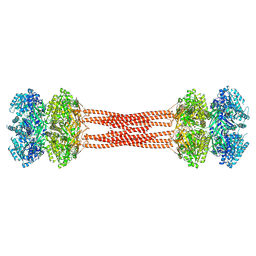 | | Crystal structure of dodecameric KaiC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Circadian clock protein KaiC, MAGNESIUM ION | | Authors: | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | Deposit date: | 2022-06-14 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
8FWI
 
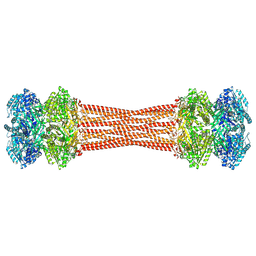 | | Structure of dodecameric KaiC-RS-S413E/S414E solved by cryo-EM | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein KaiC, ... | | Authors: | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | Deposit date: | 2023-01-22 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
8FWJ
 
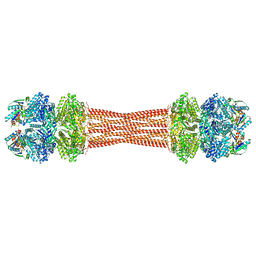 | | Structure of dodecameric KaiC-RS-S413E/S414E complexed with KaiB-RS solved by cryo-EM | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein KaiB, ... | | Authors: | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | Deposit date: | 2023-01-22 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
5FXJ
 
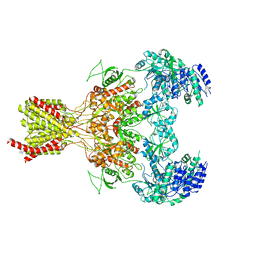 | | GluN1b-GluN2B NMDA receptor structure-Class X | | Descriptor: | GLUTAMATE RECEPTOR IONOTROPIC, NMDA 1, NMDA 2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa H, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-15 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
5FXI
 
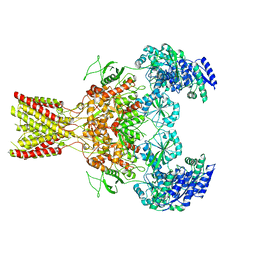 | | GluN1b-GluN2B NMDA receptor structure in non-active-2 conformation | | Descriptor: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-11 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
5FXK
 
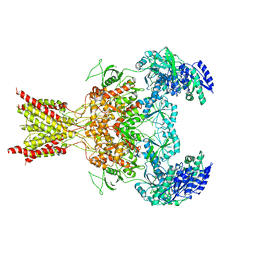 | | GluN1b-GluN2B NMDA receptor structure-Class Y | | Descriptor: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-11 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
5FXG
 
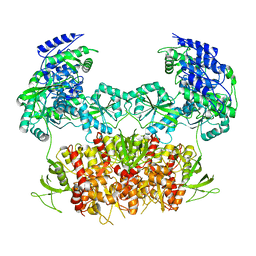 | | GLUN1B-GLUN2B NMDA RECEPTOR IN ACTIVE CONFORMATION | | Descriptor: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-11 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
5FXH
 
 | | GluN1b-GluN2B NMDA receptor in non-active-1 conformation | | Descriptor: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-11 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
5B3J
 
 | | Activation of NMDA receptors and the mechanism of inhibition by ifenprodil | | Descriptor: | Fab, heavy chain, light chain, ... | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | Deposit date: | 2016-03-01 | | Release date: | 2016-05-11 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Activation of NMDA receptors and the mechanism of inhibition by ifenprodil
Nature, 534, 2016
|
|
6MST
 
 | | Cryo-EM structure of human AA amyloid fibril | | Descriptor: | Serum amyloid A-1 protein | | Authors: | Loerch, S, Rennegarbe, M, Liberta, F, Grigorieff, N, Fandrich, M, Schmidt, M. | | Deposit date: | 2018-10-18 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM fibril structures from systemic AA amyloidosis reveal the species complementarity of pathological amyloids.
Nat Commun, 10, 2019
|
|
6AWD
 
 | | Structure of 30S (S1 depleted) ribosomal subunit and RNA polymerase complex | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
6AWB
 
 | | Structure of 30S ribosomal subunit and RNA polymerase complex in non-rotated state | | Descriptor: | 16S rRNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
6AWC
 
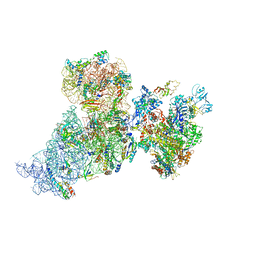 | | Structure of 30S ribosomal subunit and RNA polymerase complex in rotated state | | Descriptor: | 16S rRNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
5J0N
 
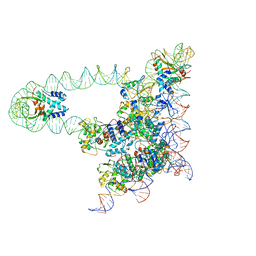 | | Lambda excision HJ intermediate | | Descriptor: | Excisionase, Integrase, Integration host factor subunit alpha, ... | | Authors: | Van Duyne, G, Grigorieff, N, Landy, A. | | Deposit date: | 2016-03-28 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Structure of a Holliday junction complex reveals mechanisms governing a highly regulated DNA transaction.
Elife, 5, 2016
|
|
4AU6
 
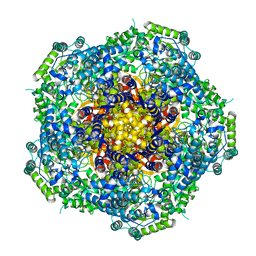 | | Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles | | Descriptor: | RNA-DEPENDENT RNA POLYMERASE | | Authors: | Estrozi, L.F, Settembre, E.C, Goret, G, McClain, B, Zhang, X, Chen, J.Z, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-13 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Location of the Dsrna-Dependent Polymerase, Vp1, in Rotavirus Particles.
J.Mol.Biol., 425, 2013
|
|
4DFC
 
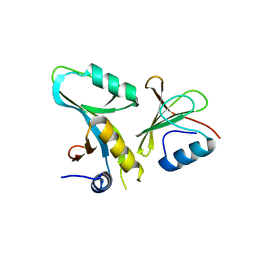 | | Core UvrA/TRCF complex | | Descriptor: | Transcription-repair-coupling factor, UvrABC system protein A | | Authors: | Deaconescu, A.M, Grigorieff, N. | | Deposit date: | 2012-01-23 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Nucleotide excision repair (NER) machinery recruitment by the transcription-repair coupling factor involves unmasking of a conserved intramolecular interface.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6U1X
 
 | | Structure of the Vesicular Stomatitis Virus L Protein in Complex with Its Phosphoprotein Cofactor (3.0 A resolution) | | Descriptor: | Phosphoprotein, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Jenni, S, Bloyet, L.M, Dias-Avalos, R, Liang, B, Wheelman, S.P.J, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2019-08-17 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the Vesicular Stomatitis Virus L Protein in Complex with Its Phosphoprotein Cofactor.
Cell Rep, 30, 2020
|
|
4F5X
 
 | | Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles | | Descriptor: | Intermediate capsid protein VP6, RNA-directed RNA polymerase, VP2 protein, ... | | Authors: | Estrozi, L.F, Settembre, E.C, Goret, G, McClain, B, Zhang, X, Chen, J.Z, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2012-05-13 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Location of the dsRNA-Dependent Polymerase, VP1, in Rotavirus Particles.
J.Mol.Biol., 425, 2013
|
|
