1B9F
 
 | | MOBILITY OF AN HIV-1 INTEGRASE ACTIVE SITE LOOP IS CORRELATED WITH CATALYTIC ACTIVITY | | Descriptor: | CACODYLATE ION, PROTEIN (INTEGRASE), SULFATE ION | | Authors: | Greenwald, J, Le, V, Butler, S.L, Bushman, F.D, Choe, S. | | Deposit date: | 1999-02-11 | | Release date: | 1999-07-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity.
Biochemistry, 38, 1999
|
|
1B92
 
 | | MOBILITY OF AN HIV-1 INTEGRASE ACTIVE SITE LOOP IS CORRELATED WITH CATALYTIC ACTIVITY | | Descriptor: | CACODYLATE ION, PROTEIN (INTEGRASE), SULFATE ION | | Authors: | Greenwald, J, Le, V, Butler, S.L, Bushman, F.D, Choe, S. | | Deposit date: | 1999-02-19 | | Release date: | 1999-07-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity.
Biochemistry, 38, 1999
|
|
1B9D
 
 | | MOBILITY OF AN HIV-1 INTEGRASE ACTIVE SITE LOOP IS CORRELATED WITH CATALYTIC ACTIVITY | | Descriptor: | CACODYLATE ION, PROTEIN (INTEGRASE), SULFATE ION | | Authors: | Greenwald, J, Le, V, Butler, S.L, Bushman, F.D, Choe, S. | | Deposit date: | 1999-02-11 | | Release date: | 1999-07-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity.
Biochemistry, 38, 1999
|
|
1BTE
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF THE TYPE II ACTIVIN RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (ACTIVIN RECEPTOR TYPE II) | | Authors: | Greenwald, J, Fischer, W, Vale, W, Choe, S. | | Deposit date: | 1998-09-01 | | Release date: | 1999-02-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-finger toxin fold for the extracellular ligand-binding domain of the type II activin receptor serine kinase.
Nat.Struct.Biol., 6, 1999
|
|
1S4Y
 
 | | Crystal structure of the activin/actrIIb extracellular domain | | Descriptor: | Activin receptor type IIB precursor, Inhibin beta A chain | | Authors: | Greenwald, J, Vega, M.E, Allendorph, G.P, Fischer, W.H, Vale, W, Choe, S, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2004-01-19 | | Release date: | 2004-08-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Flexible Activin Explains the Membrane-Dependent Cooperative Assembly of TGF-beta Family Receptors.
Mol.Cell, 15, 2004
|
|
1LX5
 
 | | Crystal Structure of the BMP7/ActRII Extracellular Domain Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin Type II Receptor, alpha-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-4)][alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Greenwald, J, Groppe, J, Kwiatkowski, W, Choe, S. | | Deposit date: | 2002-06-04 | | Release date: | 2003-04-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The BMP7/ActRII Extracellular Domain Complex Provides New Insights into
the Cooperative Nature of Receptor Assembly
Mol.Cell, 11, 2003
|
|
1LXI
 
 | | Refinement of BMP7 crystal structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BONE MORPHOGENETIC PROTEIN 7 | | Authors: | Greenwald, J, Groppe, J, Kwiatkowski, W, Choe, S. | | Deposit date: | 2002-06-05 | | Release date: | 2003-04-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The BMP7/ActRII Extracellular Domain Complex Provides New Insights into
the Cooperative Nature of Receptor Assembly
Mol.Cell, 11, 2003
|
|
2W77
 
 | | Structures of P. aeruginosa FpvA bound to heterologous pyoverdines: FpvA-Pvd(Pfl18.1)-Fe complex | | Descriptor: | (1S)-1-CARBOXY-5-[(3-CARBOXYPROPANOYL)AMINO]-8,9-DIHYDROXY-1,2,3,4-TETRAHYDROPYRIMIDO[1,2-A]QUINOLIN-11-IUM, 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, FE (III) ION, ... | | Authors: | Greenwald, J, Nader, M, Celia, H, Gruffaz, C, Meyer, J.-M, Schalk, I.J, Pattus, F. | | Deposit date: | 2008-12-20 | | Release date: | 2009-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Fpva Bound to Non-Cognate Pyoverdines: Molecular Basis of Siderophore Recognition by an Iron Transporter.
Mol.Microbiol., 72, 2009
|
|
2W75
 
 | | Structures of P. aeruginosa FpvA bound to heterologous pyoverdines: Apo-FpvA | | Descriptor: | 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, FERRIPYOVERDINE RECEPTOR, PHOSPHATE ION | | Authors: | Greenwald, J, Nader, M, Celia, H, Gruffaz, C, Meyer, J.-M, Schalk, I.J, Pattus, F. | | Deposit date: | 2008-12-20 | | Release date: | 2009-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Fpva Bound to Non-Cognate Pyoverdines: Molecular Basis of Siderophore Recognition by an Iron Transporter.
Mol.Microbiol., 72, 2009
|
|
2W16
 
 | | Structures of FpvA bound to heterologous pyoverdines | | Descriptor: | (1S)-1-CARBOXY-5-[(3-CARBOXYPROPANOYL)AMINO]-8,9-DIHYDROXY-1,2,3,4-TETRAHYDROPYRIMIDO[1,2-A]QUINOLIN-11-IUM, 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, DSN-ARG-DSN-FHO-LYS-FHO-THR-THR, ... | | Authors: | Greenwald, J, Nader, M, Celia, H, Gruffaz, C, Meyer, J.-M, Schalk, I.J, Pattus, F. | | Deposit date: | 2008-10-14 | | Release date: | 2009-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Fpva Bound to Non-Cognate Pyoverdines: Molecular Basis of Siderophore Recognition by an Iron Transporter.
Mol.Microbiol., 72, 2009
|
|
2W6U
 
 | | Structures of P. aeruginosa FpvA bound to heterologous pyoverdines: FpvA-Pvd(G173)-Fe complex | | Descriptor: | (1S)-1-CARBOXY-5-[(3-CARBOXYPROPANOYL)AMINO]-8,9-DIHYDROXY-1,2,3,4-TETRAHYDROPYRIMIDO[1,2-A]QUINOLIN-11-IUM, 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, FE (III) ION, ... | | Authors: | Greenwald, J, Nader, M, Celia, H, Gruffaz, C, Meyer, J.-M, Schalk, I.J, Pattus, F. | | Deposit date: | 2008-12-19 | | Release date: | 2009-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Fpva Bound to Non-Cognate Pyoverdines: Molecular Basis of Siderophore Recognition by an Iron Transporter.
Mol.Microbiol., 72, 2009
|
|
2W76
 
 | | Structures of P. aeruginosa FpvA bound to heterologous pyoverdines: FpvA-Pvd(Pa6)-Fe complex | | Descriptor: | (1S)-1-CARBOXY-5-[(3-CARBOXYPROPANOYL)AMINO]-8,9-DIHYDROXY-1,2,3,4-TETRAHYDROPYRIMIDO[1,2-A]QUINOLIN-11-IUM, 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, FE (III) ION, ... | | Authors: | Greenwald, J, Nader, M, Celia, H, Gruffaz, C, Meyer, J.-M, Schalk, I.J, Pattus, F. | | Deposit date: | 2008-12-20 | | Release date: | 2009-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fpva Bound to Non-Cognate Pyoverdines: Molecular Basis of Siderophore Recognition by an Iron Transporter.
Mol.Microbiol., 72, 2009
|
|
2W78
 
 | | Structures of P. aeruginosa FpvA bound to heterologous pyoverdines: FpvA-Pvd(ATCC13535)-Fe complex | | Descriptor: | (1S)-1-CARBOXY-5-[(3-CARBOXYPROPANOYL)AMINO]-8,9-DIHYDROXY-1,2,3,4-TETRAHYDROPYRIMIDO[1,2-A]QUINOLIN-11-IUM, 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, FE (III) ION, ... | | Authors: | Greenwald, J, Nader, M, Celia, H, Gruffaz, C, Meyer, J.-M, Schalk, I.J, Pattus, F. | | Deposit date: | 2008-12-20 | | Release date: | 2009-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Fpva Bound to Non-Cognate Pyoverdines: Molecular Basis of Siderophore Recognition by an Iron Transporter.
Mol.Microbiol., 72, 2009
|
|
2W6T
 
 | | Structures of P. aeruginosa FpvA bound to heterologous pyoverdines: FpvA-Pvd(DSM50106)-Fe complex | | Descriptor: | (1S)-1-CARBOXY-5-[(3-CARBOXYPROPANOYL)AMINO]-8,9-DIHYDROXY-1,2,3,4-TETRAHYDROPYRIMIDO[1,2-A]QUINOLIN-11-IUM, 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, FE (III) ION, ... | | Authors: | Greenwald, J, Nader, M, Celia, H, Gruffaz, C, Meyer, J.-M, Schalk, I.J, Pattus, F. | | Deposit date: | 2008-12-19 | | Release date: | 2009-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Fpva Bound to Non-Cognate Pyoverdines: Molecular Basis of Siderophore Recognition by an Iron Transporter.
Mol.Microbiol., 72, 2009
|
|
2WVN
 
 | | Structure of the HET-s N-terminal domain | | Descriptor: | SMALL S PROTEIN | | Authors: | Greenwald, J, Buhtz, C, Ritter, C, Kwiatkowski, W, Choe, S, Saupe, S.J, Riek, R. | | Deposit date: | 2009-10-19 | | Release date: | 2010-07-28 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The Mechanism of Prion Inhibition by Het-S.
Mol.Cell, 38, 2010
|
|
2WVQ
 
 | | Structure of the HET-s N-terminal domain. Mutant D23A, P33H | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, SMALL S PROTEIN | | Authors: | Greenwald, J, Buhtz, C, Ritter, C, Kwiatkowski, W, Choe, S, Saupe, S.J, Riek, R. | | Deposit date: | 2009-10-19 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The mechanism of prion inhibition by HET-S.
Mol. Cell, 38, 2010
|
|
2WVO
 
 | | Structure of the HET-S N-terminal domain | | Descriptor: | CHLORIDE ION, SMALL S PROTEIN | | Authors: | Greenwald, J, Buhtz, C, Ritter, C, Kwiatkowski, W, Choe, S, Saupe, S.J, Riek, R. | | Deposit date: | 2009-10-19 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Mechanism of Prion Inhibition by Het-S.
Mol.Cell, 38, 2010
|
|
8PXS
 
 | | Short RNA binding to peptide amyloids | | Descriptor: | RNA (5'-R(P*GP*UP*CP*A)-3'), VAL-ALA-GLN-ALA-GLN-ILE-ASN-ILE | | Authors: | Rout, S.K, Cadalbert, R, Schroder, N, Wiegand, T, Zehnder, J, Gampp, O, Guntert, P, Kringler, D, Kreutz, C, Knorlein, A, Hall, J, Greenwald, J, Riek, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-10-18 | | Method: | SOLID-STATE NMR | | Cite: | An Analysis of Nucleotide-Amyloid Interactions Reveals Selective Binding to Codon-Sized RNA.
J.Am.Chem.Soc., 145, 2023
|
|
1YGM
 
 | | NMR structure of Mistic | | Descriptor: | hypothetical protein BSU31320 | | Authors: | Roosild, T.P, Greenwald, J, Vega, M, Castronovo, S, Riek, R, Choe, S. | | Deposit date: | 2005-01-05 | | Release date: | 2005-03-01 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Mistic, a membrane-integrating protein for membrane protein expression.
Science, 307, 2005
|
|
3OTX
 
 | | Crystal Structure of Trypanosoma brucei rhodesiense Adenosine Kinase Complexed with Inhibitor AP5A | | Descriptor: | Adenosine kinase, putative, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ... | | Authors: | Kuettel, S, Greenwald, J, Kostrewa, D, Ahmed, S, Scapozza, L, Perozzo, R. | | Deposit date: | 2010-09-14 | | Release date: | 2011-06-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structures of T. b. rhodesiense Adenosine Kinase Complexed with Inhibitor and Activator: Implications for Catalysis and Hyperactivation
Plos Negl Trop Dis, 5, 2011
|
|
1M4U
 
 | | Crystal structure of Bone Morphogenetic Protein-7 (BMP-7) in complex with the secreted antagonist Noggin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bone Morphogenetic Protein-7, Noggin | | Authors: | Groppe, J, Greenwald, J, Wiater, E, Rodriguez-Leon, J, Economides, A.N, Kwiatkowski, W, Affolter, M, Vale, W.W, Izpisua-Belmonte, J.C, Choe, S. | | Deposit date: | 2002-07-03 | | Release date: | 2002-12-18 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Basis of BMP Signalling Inhibition by the Cystine Knot Protein Noggin
Nature, 420, 2002
|
|
1HYV
 
 | | HIV INTEGRASE CORE DOMAIN COMPLEXED WITH TETRAPHENYL ARSONIUM | | Descriptor: | CHLORIDE ION, INTEGRASE, SULFATE ION, ... | | Authors: | Molteni, V, Greenwald, J, Rhodes, D, Hwang, Y, Kwiatkowski, W, Bushman, F.D, Siegel, J.S, Choe, S. | | Deposit date: | 2001-01-22 | | Release date: | 2001-04-04 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of a small-molecule binding site at the dimer interface of the HIV integrase catalytic domain.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HYZ
 
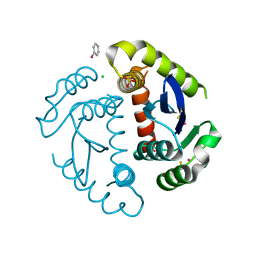 | | HIV INTEGRASE CORE DOMAIN COMPLEXED WITH A DERIVATIVE OF TETRAPHENYL ARSONIUM. | | Descriptor: | (3,4-DIHYDROXY-PHENYL)-TRIPHENYL-ARSONIUM, CHLORIDE ION, INTEGRASE, ... | | Authors: | Molteni, V, Greenwald, J, Rhodes, D, Hwang, Y, Kwiatkowski, W, Bushman, F.D, Siegel, J.S, Choe, S. | | Deposit date: | 2001-01-22 | | Release date: | 2001-04-04 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of a small-molecule binding site at the dimer interface of the HIV integrase catalytic domain.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2XTB
 
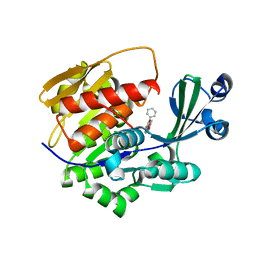 | | Crystal Structure of Trypanosoma brucei rhodesiense Adenosine Kinase Complexed with Activator | | Descriptor: | 4-[5-(4-PHENOXYPHENYL)-1H-PYRAZOL-3-YL]MORPHOLINE, ADENOSINE KINASE | | Authors: | Kuettel, S, Greenwald, J, Kostrewa, D, Ahmed, S, Scapozza, L, Perozzo, R. | | Deposit date: | 2010-10-05 | | Release date: | 2011-06-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of T. B. Rhodesiense Adenosine Kinase Complexed with Inhibitor and Activator: Implications for Catalysis and Hyperactivation.
Plos Negl Trop Dis, 5, 2011
|
|
8TJY
 
 | | Structure of Gabija AB complex | | Descriptor: | Endonuclease GajA, Gabija protein GajB | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-07-24 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Molecular basis of Gabija anti-phage supramolecular assemblies
Nat.Struct.Mol.Biol., 2024
|
|
