1CSZ
 
 | | SYK TYROSINE KINASE C-TERMINAL SH2 DOMAIN COMPLEXED WITH A PHOSPHOPEPTIDEFROM THE GAMMA CHAIN OF THE HIGH AFFINITY IMMUNOGLOBIN G RECEPTOR, NMR | | Descriptor: | ACETYL-THR-PTR-GLU-THR-LEU-NH2, SYK PROTEIN TYROSINE KINASE | | Authors: | Narula, S.S, Yuan, R.W, Adams, S.E, Green, O.M, Green, J, Phillips, T.B, Zydowsky, L.D, Botfield, M.C, Hatada, M.H, Laird, E.R, Zoller, M.J, Karas, J.L, Dalgarno, D.C. | | Deposit date: | 1995-10-03 | | Release date: | 1996-11-08 | | Last modified: | 2012-02-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal SH2 domain of the human tyrosine kinase Syk complexed with a phosphotyrosine pentapeptide.
Structure, 3, 1995
|
|
1CSY
 
 | | SYK TYROSINE KINASE C-TERMINAL SH2 DOMAIN COMPLEXED WITH A PHOSPHOPEPTIDEFROM THE GAMMA CHAIN OF THE HIGH AFFINITY IMMUNOGLOBIN G RECEPTOR, NMR | | Descriptor: | ACETYL-THR-PTR-GLU-THR-LEU-NH2, SYK PROTEIN TYROSINE KINASE | | Authors: | Narula, S.S, Yuan, R.W, Adams, S.E, Green, O.M, Green, J, Phillips, T.B, Zydowsky, L.D, Botfield, M.C, Hatada, M.H, Laird, E.R, Zoller, M.J, Karas, J.L, Dalgarno, D.C. | | Deposit date: | 1995-10-03 | | Release date: | 1996-11-08 | | Last modified: | 2012-02-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal SH2 domain of the human tyrosine kinase Syk complexed with a phosphotyrosine pentapeptide.
Structure, 3, 1995
|
|
7HVP
 
 | | X-RAY CRYSTALLOGRAPHIC STRUCTURE OF A COMPLEX BETWEEN A SYNTHETIC PROTEASE OF HUMAN IMMUNODEFICIENCY VIRUS 1 AND A SUBSTRATE-BASED HYDROXYETHYLAMINE INHIBITOR | | Descriptor: | HIV-1 PROTEASE, INHIBITOR ACE-SER-LEU-ASN-PHE-PSI(CH(OH)-CH2N)-PRO-ILE VME (JG-365) | | Authors: | Swain, A.L, Miller, M.M, Green, J, Rich, D.H, Schneider, J, Kent, S.B.H, Wlodawer, A. | | Deposit date: | 1990-09-13 | | Release date: | 1993-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic structure of a complex between a synthetic protease of human immunodeficiency virus 1 and a substrate-based hydroxyethylamine inhibitor.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
5LBM
 
 | | The asymmetric tetrameric structure of the formaldehyde sensing transcriptional repressor FrmR from Escherichia coli | | Descriptor: | FORMYL GROUP, Transcriptional repressor FrmR | | Authors: | Bisson, C, Baker, P.J, Green, J, Chivers, P.T. | | Deposit date: | 2016-06-16 | | Release date: | 2016-12-21 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The mechanism of a formaldehyde-sensing transcriptional regulator.
Sci Rep, 6, 2016
|
|
1ZUJ
 
 | | The crystal structure of the Lactococcus lactis MG1363 DpsA protein | | Descriptor: | hypothetical protein Llacc01001955 | | Authors: | Stillman, T.J, Upadhyay, M, Norte, V.A, Sedelnikova, S.E, Carradus, M, Tzokov, S, Bullough, P.A, Shearman, C.A, Gasson, M.J, Williams, C.H, Artymiuk, P.J, Green, J. | | Deposit date: | 2005-05-31 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structures of Lactococcus lactis MG1363 Dps proteins reveal the presence of an N-terminal helix that is required for DNA binding.
Mol.Microbiol., 57, 2005
|
|
1ZS3
 
 | | The crystal structure of the Lactococcus lactis MG1363 DpsB protein | | Descriptor: | Lactococcus lactis MG1363 DpsA | | Authors: | Stillman, T.J, Upadhyay, M, Norte, V.A, Sedelnikova, S.E, Carradus, M, Tzokov, S, Bullough, P.A, Shearman, C.A, Gasson, M.J, Williams, C.H, Artymiuk, P.J, Green, J. | | Deposit date: | 2005-05-23 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structures of Lactococcus lactis MG1363 Dps proteins reveal the presence of an N-terminal helix that is required for DNA binding.
Mol.Microbiol., 57, 2005
|
|
5OAY
 
 | |
2OQ1
 
 | | Tandem SH2 domains of ZAP-70 with 19-mer zeta1 peptide | | Descriptor: | LEAD (II) ION, T-cell surface glycoprotein CD3 zeta chain, Tyrosine-protein kinase ZAP-70 | | Authors: | Hatada, M.H, Laird, E.R, Green, J, Morgenstern, J, Ram, M.K. | | Deposit date: | 2007-01-30 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for the interaction of ZAP-70 with the T-cell receptor
Nature, 377, 1995
|
|
2OT5
 
 | |
1QOY
 
 | | E.coli Hemolysin E (HlyE, ClyA, SheA) | | Descriptor: | HEMOLYSIN E, SULFATE ION | | Authors: | Wallace, A.J, Stillman, T.J, Atkins, A, Jamieson, S.J, Bullough, P.A, Green, J, Artymiuk, P.J. | | Deposit date: | 1999-11-25 | | Release date: | 2000-01-23 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | E. Coli Hemolysin E (Hlye, Clya, Shea): X-Ray Crystal Structure of the Toxin and Observation of Membrane Pores by Electron Microscopy
Cell(Cambridge,Mass.), 100, 2000
|
|
1L5J
 
 | | CRYSTAL STRUCTURE OF E. COLI ACONITASE B. | | Descriptor: | ACONITATE ION, Aconitate hydratase 2, FE3-S4 CLUSTER | | Authors: | Williams, C.H, Stillman, T.J, Barynin, V.V, Sedelnikova, S.E, Tang, Y, Green, J, Guest, J.R, Artymiuk, P.J. | | Deposit date: | 2002-03-07 | | Release date: | 2002-06-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | E. coli aconitase B structure reveals a HEAT-like domain with implications for protein-protein recognition.
Nat.Struct.Biol., 9, 2002
|
|
4YVE
 
 | | ROCK 1 bound to methoxyphenyl thiazole inhibitor | | Descriptor: | 2-(3-methoxyphenyl)-N-[4-(pyridin-4-yl)-1,3-thiazol-2-yl]acetamide, Rho-associated protein kinase 1 | | Authors: | Jacobs, M.D. | | Deposit date: | 2015-03-19 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Design, Synthesis, and Structure-Activity Relationships of Pyridine-Based Rho Kinase (ROCK) Inhibitors.
J.Med.Chem., 58, 2015
|
|
4YVC
 
 | | ROCK 1 bound to thiazole inhibitor | | Descriptor: | 2-fluoro-N-[4-(pyridin-4-yl)-1,3-thiazol-2-yl]benzamide, Rho-associated protein kinase 1 | | Authors: | Jacobs, M.D. | | Deposit date: | 2015-03-19 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design, Synthesis, and Structure-Activity Relationships of Pyridine-Based Rho Kinase (ROCK) Inhibitors.
J.Med.Chem., 58, 2015
|
|
5BML
 
 | | ROCK 1 bound to a pyridine thiazole inhibitor | | Descriptor: | N-[4-(2-fluoropyridin-4-yl)thiophen-2-yl]-2-{3-[(methylsulfonyl)amino]phenyl}acetamide, Rho-associated protein kinase 1 | | Authors: | Jacobs, M.D. | | Deposit date: | 2015-05-22 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Design, Synthesis, and Structure-Activity Relationships of Pyridine-Based Rho Kinase (ROCK) Inhibitors.
J.Med.Chem., 58, 2015
|
|
7RCU
 
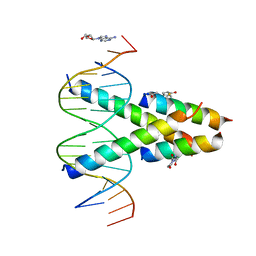 | | Synthetic Max homodimer mimic in complex with DNA | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 2-(2,5-dioxopyrrolidin-1-yl)acetamide, ACETAMIDE, ... | | Authors: | Speltz, T, Qiao, Z, Shangguan, S, Fanning, S, Greene, J, Moellering, R. | | Deposit date: | 2021-07-08 | | Release date: | 2022-09-14 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Targeting MYC with modular synthetic transcriptional repressors derived from bHLH DNA-binding domains.
Nat.Biotechnol., 41, 2023
|
|
1CR6
 
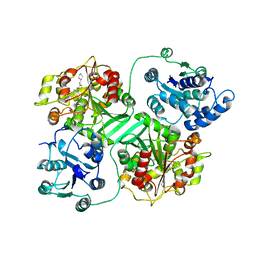 | | CRYSTAL STRUCTURE OF MURINE SOLUBLE EPOXIDE HYDROLASE COMPLEXED WITH CPU INHIBITOR | | Descriptor: | EPOXIDE HYDROLASE, N-CYCLOHEXYL-N'-(PROPYL)PHENYL UREA | | Authors: | Argiriadi, M.A, Morisseau, C, Hammock, B.D, Christianson, D.W. | | Deposit date: | 1999-08-13 | | Release date: | 1999-11-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Detoxification of environmental mutagens and carcinogens: structure, mechanism, and evolution of liver epoxide hydrolase.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
5HVU
 
 | |
3I60
 
 | |
3I5Z
 
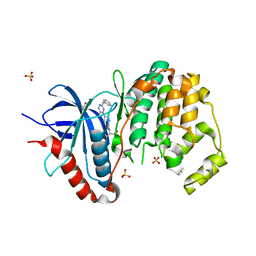 | |
3I4B
 
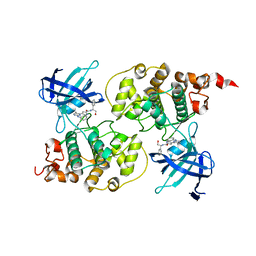 | | Crystal structure of GSK3b in complex with a pyrimidylpyrrole inhibitor | | Descriptor: | Glycogen synthase kinase-3 beta, N-[(1S)-2-hydroxy-1-phenylethyl]-4-[5-methyl-2-(phenylamino)pyrimidin-4-yl]-1H-pyrrole-2-carboxamide | | Authors: | Ter Haar, E. | | Deposit date: | 2009-07-01 | | Release date: | 2010-01-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided design of potent and selective pyrimidylpyrrole inhibitors of extracellular signal-regulated kinase (ERK) using conformational control.
J.Med.Chem., 52, 2009
|
|
5KKT
 
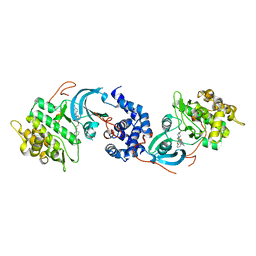 | | ROCK 1 bound to azaindole thiazole piperazine inhibitor | | Descriptor: | 2-[3-[3-(4-methylpiperazin-1-yl)propoxy]phenyl]-~{N}-[4-(1~{H}-pyrrolo[2,3-b]pyridin-3-yl)-1,3-thiazol-2-yl]ethanamide, Rho-associated protein kinase 1 | | Authors: | Jacobs, M.D. | | Deposit date: | 2016-06-22 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | ROCK inhibitors 3: Design, synthesis and structure-activity relationships of 7-azaindole-based Rho kinase (ROCK) inhibitors
Bioorg.Med.Chem.Lett., 2018
|
|
5KKS
 
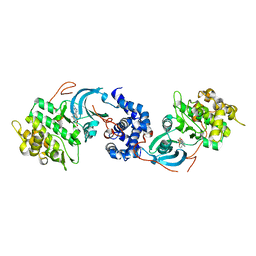 | | ROCK 1 bound to azaindole thiazole inhibitor | | Descriptor: | 2-[3-(methylsulfonylamino)phenyl]-~{N}-[4-(1~{H}-pyrrolo[2,3-b]pyridin-3-yl)-1,3-thiazol-2-yl]ethanamide, Rho-associated protein kinase 1 | | Authors: | Jacobs, M.D. | | Deposit date: | 2016-06-22 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | ROCK inhibitors 3: Design, synthesis and structure-activity relationships of 7-azaindole-based Rho kinase (ROCK) inhibitors
Bioorg.Med.Chem.Lett., 2018
|
|
5UZK
 
 | | Crystal Structure of PKA bound to an pyrrolo pyridine inhibitor | | Descriptor: | 2-{3-[3-(piperidin-4-yl)propoxy]phenyl}-N-[4-(1H-pyrrolo[2,3-b]pyridin-3-yl)-1,3-thiazol-2-yl]acetamide, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Jacobs, M.D, Brown, K. | | Deposit date: | 2017-02-26 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ROCK inhibitors 3: Design, synthesis and structure-activity relationships of 7-azaindole-based Rho kinase (ROCK) inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
5UZJ
 
 | | Crystal Structure of ROCK1 bound to an aminopyridine inhibitor | | Descriptor: | N-[4-(2-aminopyridin-4-yl)-1,3-thiazol-2-yl]-2-(3-methoxyphenyl)acetamide, Rho-associated protein kinase 1 | | Authors: | Jacobs, M.D. | | Deposit date: | 2017-02-26 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | ROCK inhibitors 3: Design, synthesis and structure-activity relationships of 7-azaindole-based Rho kinase (ROCK) inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
3L1S
 
 | |
